Gene body DNA hydroxymethylation restricts the magnitude of transcriptional changes during aging
- PMID: 39069555
- PMCID: PMC11284234
- DOI: 10.1038/s41467-024-50725-y
Gene body DNA hydroxymethylation restricts the magnitude of transcriptional changes during aging
Abstract
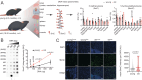
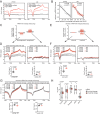
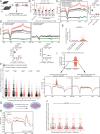
DNA hydroxymethylation (5hmC), the most abundant oxidative derivative of DNA methylation, is typically enriched at enhancers and gene bodies of transcriptionally active and tissue-specific genes. Although aberrant genomic 5hmC has been implicated in age-related diseases, its functional role in aging remains unknown. Here, using mouse liver and cerebellum as model organs, we show that 5hmC accumulates in gene bodies associated with tissue-specific function and restricts the magnitude of gene expression changes with age. Mechanistically, 5hmC decreases the binding of splicing associated factors and correlates with age-related alternative splicing events. We found that various age-related contexts, such as prolonged quiescence and senescence, drive the accumulation of 5hmC with age. We provide evidence that this age-related transcriptionally restrictive function is conserved in mouse and human tissues. Our findings reveal that 5hmC regulates tissue-specific function and may play a role in longevity.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Gene body DNA hydroxymethylation restricts the magnitude of transcriptional changes during aging.bioRxiv [Preprint]. 2024 Jul 3:2023.02.15.528714. doi: 10.1101/2023.02.15.528714. bioRxiv. 2024. Update in: Nat Commun. 2024 Jul 28;15(1):6357. doi: 10.1038/s41467-024-50725-y PMID: 36824863 Free PMC article. Updated. Preprint.
Similar articles
-
Correlated 5-Hydroxymethylcytosine (5hmC) and Gene Expression Profiles Underpin Gene and Organ-Specific Epigenetic Regulation in Adult Mouse Brain and Liver.PLoS One. 2017 Jan 26;12(1):e0170779. doi: 10.1371/journal.pone.0170779. eCollection 2017. PLoS One. 2017. PMID: 28125731 Free PMC article.
-
Analysis of the machinery and intermediates of the 5hmC-mediated DNA demethylation pathway in aging on samples from the MARK-AGE Study.Aging (Albany NY). 2016 Aug 29;8(9):1896-1922. doi: 10.18632/aging.101022. Aging (Albany NY). 2016. PMID: 27587280 Free PMC article.
-
Genome-wide characterization of cytosine-specific 5-hydroxymethylation in normal breast tissue.Epigenetics. 2020 Apr;15(4):398-418. doi: 10.1080/15592294.2019.1695332. Epub 2019 Dec 16. Epigenetics. 2020. PMID: 31842685 Free PMC article.
-
Environmental Epigenetics and Genome Flexibility: Focus on 5-Hydroxymethylcytosine.Int J Mol Sci. 2020 May 2;21(9):3223. doi: 10.3390/ijms21093223. Int J Mol Sci. 2020. PMID: 32370155 Free PMC article. Review.
-
The elusive role of 5'-hydroxymethylcytosine.Epigenomics. 2016 Nov;8(11):1539-1551. doi: 10.2217/epi-2016-0076. Epub 2016 Oct 13. Epigenomics. 2016. PMID: 27733056 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

