First report of fesavirus 4 detection from cats in Japan
- PMID: 39069477
- PMCID: PMC11422691
- DOI: 10.1292/jvms.24-0243
First report of fesavirus 4 detection from cats in Japan
Abstract
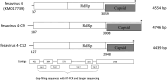
Fesaviruses, picorna-like RNA viruses, were discovered in 2014 in feces from cats in an animal shelter in the United States but have not since been reported elsewhere. In this study, we collected cat fecal samples from 20 adult cats from an animal shelter in Tokyo, Japan, and examined them for viral pathogens. Next generation sequencing (NGS) was performed to detect both RNA and DNA virus sequences. Sequences of a total of 7 RNA viruses including some common feline pathogenic viruses were detected across 8 samples, while no DNA virus sequences were identified in any sample. Of the RNA virus sequences detected in the samples, two sequences, 4,746 and 4,439 bp, demonstrated 90.3% and 85.0% similarity, respectively, to the fesavirus 4 sequence in the database. To confirm the NGS results, quantitative RT-PCR (qRT-PCR) assays were developed using specific primers and probes designed based on the contig sequences. Based on the qRT-PCR assays, we detected relatively high copy-numbers of fesavirus 4 RNA in the two fecal samples from which the fesavirus 4 sequences were originally obtained, and low copy numbers in other samples. These results demonstrate the presence of fesavirus 4 in cats in Japan for the first time.
Keywords: animal shelter; cat feces sample; fesavirus 4; next-generation sequencing; picorna-like virus.
Conflict of interest statement
The authors declare that there is no conflict of interest involved in this study.
Figures

Similar articles
-
Identification and complete genome analysis of a novel bovine picornavirus in Japan.Virus Res. 2015 Dec 2;210:205-12. doi: 10.1016/j.virusres.2015.08.001. Epub 2015 Aug 7. Virus Res. 2015. PMID: 26260333 Free PMC article.
-
A novel sapelovirus-like virus isolation from wild boar.Virus Genes. 2011 Oct;43(2):243-8. doi: 10.1007/s11262-011-0628-2. Epub 2011 Jun 4. Virus Genes. 2011. PMID: 21643767
-
Identification of boosepivirus B in U.S. calves.Arch Virol. 2021 Nov;166(11):3193-3197. doi: 10.1007/s00705-021-05231-7. Epub 2021 Sep 15. Arch Virol. 2021. PMID: 34528138 Free PMC article.
-
Identification of a novel feline picornavirus from the domestic cat.J Virol. 2012 Jan;86(1):395-405. doi: 10.1128/JVI.06253-11. Epub 2011 Oct 26. J Virol. 2012. PMID: 22031936 Free PMC article.
-
Saliviruses-the first knowledge about a newly discovered human picornavirus.Rev Med Virol. 2017 Jan;27(1). doi: 10.1002/rmv.1904. Epub 2016 Sep 19. Rev Med Virol. 2017. PMID: 27641729 Review.
References
-
- Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, García-Cobos S, Kooistra-Smid AM, Raangs EC, Rosema S, Veloo AC, Zhou K, Friedrich AW, Rossen JW. 2017. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol 243: 16–24. doi: 10.1016/j.jbiotec.2016.12.022 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

