Increased copy-number variant load of associated risk genes in sporadic cases of amyotrophic lateral sclerosis
- PMID: 39066921
- PMCID: PMC11335238
- DOI: 10.1007/s00018-024-05335-8
Increased copy-number variant load of associated risk genes in sporadic cases of amyotrophic lateral sclerosis
Abstract
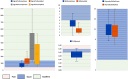
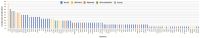
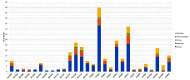
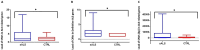
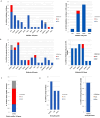
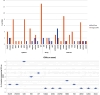
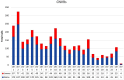
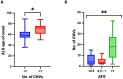
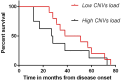
Amyotrophic lateral sclerosis (ALS) is an age-related neurodegenerative disease characterized by selective loss of motor neurons in the brainstem and spinal cord. Several genetic factors have been associated to ALS, ranging from causal genes and potential risk factors to disease modifiers. The search for pathogenic variants in these genes has mostly focused on single nucleotide variants (SNVs) while relatively understudied and not fully elucidated is the contribution of structural variants, such as copy number variations (CNVs). Here, we applied an exon-centric aCGH method to investigate, in sporadic ALS patients, the load of CNVs in 131 genes previously associated to ALS. Our approach revealed that CNV load, defined as the total number of CNVs or their size, was significantly higher in ALS cases than controls. About 87% of patients harbored multiple CNVs in ALS-related genes, and 75% structural variants compromised genes directly implicated in ALS pathogenesis (C9orf72, CHCHD10, EPHA4, FUS, HNRNPA1, KIF5A, NEK1, OPTN, PFN1, SOD1, TARDBP, TBK1, UBQLN2, UNC13A, VAPB, VCP). CNV load was also associated to higher onset age and disease progression rate. Although the contribution of individual CNVs in ALS is still unknown, their extensive load in disease-related genes may have relevant implications for the diagnostic, prognostic and therapeutical management of this devastating disorder.
Keywords: Amyotrophic lateral sclerosis; CNVs load; Copy number variant; Customized aCGH; Diagnostics.
© 2024. The Author(s).
Conflict of interest statement
Authors declare that there are any competing interests in relation to the work described.
Figures
Similar articles
-
The role of copy number variation in susceptibility to amyotrophic lateral sclerosis: genome-wide association study and comparison with published loci.PLoS One. 2009 Dec 4;4(12):e8175. doi: 10.1371/journal.pone.0008175. PLoS One. 2009. PMID: 19997636 Free PMC article.
-
Genetic variability in sporadic amyotrophic lateral sclerosis.Brain. 2023 Sep 1;146(9):3760-3769. doi: 10.1093/brain/awad120. Brain. 2023. PMID: 37043475 Free PMC article.
-
Amyotrophic Lateral Sclerosis Overview.2001 Mar 23 [updated 2023 Sep 28]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2001 Mar 23 [updated 2023 Sep 28]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301623 Free Books & Documents. Review.
-
Copy Number Variations in Amyotrophic Lateral Sclerosis: Piecing the Mosaic Tiles Together through a Systems Biology Approach.Mol Neurobiol. 2018 Feb;55(2):1299-1322. doi: 10.1007/s12035-017-0393-x. Epub 2017 Jan 24. Mol Neurobiol. 2018. PMID: 28120152 Free PMC article. Review.
-
Copy-number variation in sporadic amyotrophic lateral sclerosis: a genome-wide screen.Lancet Neurol. 2008 Apr;7(4):319-26. doi: 10.1016/S1474-4422(08)70048-6. Epub 2008 Mar 3. Lancet Neurol. 2008. PMID: 18313986
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

