Construction and Characterization of a High-Capacity Replication-Competent Murine Cytomegalovirus Vector for Gene Delivery
- PMID: 39066429
- PMCID: PMC11281640
- DOI: 10.3390/vaccines12070791
Construction and Characterization of a High-Capacity Replication-Competent Murine Cytomegalovirus Vector for Gene Delivery
Abstract
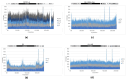
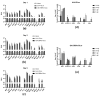
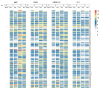
We investigated the basic characteristics of a new murine cytomegalovirus (MCMV) vector platform. Using BAC technology, we engineered replication-competent recombinant MCMVs with deletions of up to 26% of the wild-type genome. To this end, we targeted five gene blocks (m01-m17, m106-m109, m129-m141, m144-m158, and m159-m170). BACs featuring deletions from 18% to 26% of the wild-type genome exhibited delayed virus reconstitution, while smaller deletions (up to 16%) demonstrated reconstitution kinetics similar to those of the wild type. Utilizing an innovative methodology, we introduced large genomic DNA segments, up to 35 kbp, along with reporter genes into a newly designed vector with a potential cloning capacity of 46 kbp (Q4). Surprisingly, the insertion of diverse foreign DNAs alleviated the delayed plaque formation phenotype of Q4, and these large inserts remained stable through serial in vitro passages. With reporter-gene-expressing recombinant MCMVs, we successfully transduced not only mouse cell lines but also non-rodent mammalian cells, including those of human, monkey, bovine, and bat origin. Remarkably, even non-mammalian cell lines derived from chickens exhibited successful transduction.
Keywords: cross-species application; cytomegalovirus; gene transfer vector; genetic stability; high-capacity vector; host range.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Repair of an Attenuated Low-Passage Murine Cytomegalovirus Bacterial Artificial Chromosome Identifies a Novel Spliced Gene Essential for Salivary Gland Tropism.J Virol. 2020 Oct 27;94(22):e01456-20. doi: 10.1128/JVI.01456-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32847854 Free PMC article.
-
Generation of mutant murine cytomegalovirus strains from overlapping cosmid and plasmid clones.J Virol. 2000 Oct;74(19):8972-9. doi: 10.1128/jvi.74.19.8972-8979.2000. J Virol. 2000. PMID: 10982341 Free PMC article.
-
Mouse Cytomegalovirus M34 Encodes a Non-essential, Nuclear, Early-Late Expressed Protein Required for Efficient Viral Replication.Front Cell Infect Microbiol. 2020 May 5;10:171. doi: 10.3389/fcimb.2020.00171. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32432049 Free PMC article.
-
Mouse cytomegalovirus in developing brain tissue: analysis of 11 species with GFP-expressing recombinant virus.J Comp Neurol. 2000 Nov 27;427(4):559-80. doi: 10.1002/1096-9861(20001127)427:4<559::aid-cne5>3.0.co;2-4. J Comp Neurol. 2000. PMID: 11056464
-
Exploring the Potential of Cytomegalovirus-Based Vectors: A Review.Viruses. 2023 Oct 2;15(10):2043. doi: 10.3390/v15102043. Viruses. 2023. PMID: 37896820 Free PMC article. Review.
References
-
- Hansen S.G., Womack J., Scholz I., Renner A., Edgel K.A., Xu G., Ford J.C., Grey M., St Laurent B., Turner J.M., et al. Cytomegalovirus vectors expressing Plasmodium knowlesi antigens induce immune responses that delay parasitemia upon sporozoite challenge. PLoS ONE. 2019;14:e0210252. doi: 10.1371/journal.pone.0210252. - DOI - PMC - PubMed
-
- Marzi A., Murphy A.A., Feldmann F., Parkins C.J., Haddock E., Hanley P.W., Emery M.J., Engelmann F., Messaoudi I., Feldmann H., et al. Cytomegalovirus-based vaccine expressing Ebola virus glycoprotein protects nonhuman primates from Ebola virus infection. Sci. Rep. 2016;6:21674. doi: 10.1038/srep21674. - DOI - PMC - PubMed
-
- Tsuda Y., Caposio P., Parkins C.J., Botto S., Messaoudi I., Cicin-Sain L., Feldmann H., Jarvis M.A. A replicating cytomegalovirus-based vaccine encoding a single Ebola virus nucleoprotein CTL epitope confers protection against Ebola virus. PLoS Negl. Trop. Dis. 2011;5:e1275. doi: 10.1371/journal.pntd.0001275. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

