Regulation of Mitochondria-Derived Immune Activation by 'Antiviral' TRIM Proteins
- PMID: 39066323
- PMCID: PMC11281404
- DOI: 10.3390/v16071161
Regulation of Mitochondria-Derived Immune Activation by 'Antiviral' TRIM Proteins
Abstract
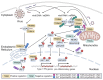
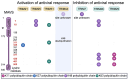
Mitochondria are key orchestrators of antiviral responses that serve as platforms for the assembly and activation of innate immune-signaling complexes. In response to viral infection, mitochondria can be triggered to release immune-stimulatory molecules that can boost interferon production. These same molecules can be released by damaged mitochondria to induce pathogenic, antiviral-like immune responses in the absence of infection. This review explores how members of the tripartite motif-containing (TRIM) protein family, which are recognized for their roles in antiviral defense, regulate mitochondria-based innate immune activation. In antiviral defense, TRIMs are essential components of immune signal transduction pathways and function as directly acting viral restriction factors. TRIMs carry out conceptually similar activities when controlling immune activation related to mitochondria. First, they modulate immune-signaling pathways that can be activated by mitochondrial molecules. Second, they co-ordinate the direct removal of mitochondria and associated immune-activating factors through mitophagy. These insights broaden the scope of TRIM actions in innate immunity and may implicate TRIMs in diseases associated with mitochondria-derived inflammation.
Keywords: MAVS; MDA5; RIG-I; STING; TBK1; TRIM; antiviral defense; autophagy; cGAS; inflammation; interferon; mitochondria; mitophagy; restriction factor; tripartite motif.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
TRIM Proteins and Their Roles in Antiviral Host Defenses.Annu Rev Virol. 2018 Sep 29;5(1):385-405. doi: 10.1146/annurev-virology-092917-043323. Epub 2018 Jun 27. Annu Rev Virol. 2018. PMID: 29949725 Free PMC article. Review.
-
Tripartite motif-containing proteins precisely and positively affect host antiviral immune response.Scand J Immunol. 2018 Jun;87(6):e12669. doi: 10.1111/sji.12669. Scand J Immunol. 2018. PMID: 29706026 Review.
-
Mechanisms and pathways of innate immune activation and regulation in health and cancer.Hum Vaccin Immunother. 2014;10(11):3270-85. doi: 10.4161/21645515.2014.979640. Hum Vaccin Immunother. 2014. PMID: 25625930 Free PMC article. Review.
-
The Roles of TRIMs in Antiviral Innate Immune Signaling.Front Cell Infect Microbiol. 2021 Mar 15;11:628275. doi: 10.3389/fcimb.2021.628275. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33791238 Free PMC article. Review.
-
Emerging views of mitophagy in immunity and autoimmune diseases.Autophagy. 2020 Jan;16(1):3-17. doi: 10.1080/15548627.2019.1603547. Epub 2019 Apr 21. Autophagy. 2020. PMID: 30951392 Free PMC article.
References
-
- Chauhan S., Kumar S., Jain A., Ponpuak M., Mudd M.H., Kimura T., Choi S.W., Peters R., Mandell M., Bruun J.A., et al. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-direct Autophagy in Endomembrane Damage Homeostasis. Dev. Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. - DOI - PMC - PubMed
-
- Romagnoli A., Di Rienzo M., Petruccioli E., Fusco C., Palucci I., Micale L., Mazza T., Delogu G., Merla G., Goletti D., et al. The ubiquitin ligase TRIM32 promotes the autophagic response to Mycobacterium tuberculosis infection in macrophages. Cell Death Dis. 2023;14:505. doi: 10.1038/s41419-023-06026-1. - DOI - PMC - PubMed
-
- Hoffpauir C.T., Bell S.L., West K.O., Jing T., Wagner A.R., Torres-Odio S., Cox J.S., West A.P., Li P., Patrick K.L., et al. TRIM14 Is a Key Regulator of the Type I IFN Response during Mycobacterium tuberculosis Infection. J. Immunol. 2020;205:153–167. doi: 10.4049/jimmunol.1901511. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

