Primed for Interactions: Investigating the Primed Substrate Channel of the Proteasome for Improved Molecular Engagement
- PMID: 39064934
- PMCID: PMC11279888
- DOI: 10.3390/molecules29143356
Primed for Interactions: Investigating the Primed Substrate Channel of the Proteasome for Improved Molecular Engagement
Abstract
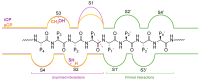
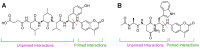
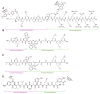
Protein homeostasis is a tightly conserved process that is regulated through the ubiquitin proteasome system (UPS) in a ubiquitin-independent or ubiquitin-dependent manner. Over the past two decades, the proteasome has become an excellent therapeutic target through inhibition of the catalytic core particle, inhibition of subunits responsible for recognizing and binding ubiquitinated proteins, and more recently, through targeted protein degradation using proteolysis targeting chimeras (PROTACs). The majority of the developed inhibitors of the proteasome's core particle rely on gaining selectivity through binding interactions within the unprimed substrate channel. Although this has allowed for selective inhibitors and chemical probes to be generated for the different proteasome isoforms, much remains unknown about the interactions that could be harnessed within the primed substrate channel to increase potency or selectivity. Herein, we discuss small molecules that interact with the primed substrate pocket and how their differences may give rise to altered activity. Taking advantage of additional interactions with the primed substrate pocket of the proteasome could allow for the generation of improved chemical tools for perturbing or monitoring proteasome activity.
Keywords: inhibitor; proteasome; substrate channel.
Conflict of interest statement
Prof. Trader is a shareholder and consultant for Booster Therapeutics, GmbH. Other authors declares no conflicts of interest.
Figures




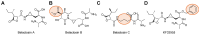
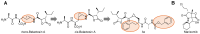
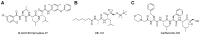
Similar articles
-
20S proteasome hydrolysis of LLVY substrates to determine preferences for moieties in its primed substrate channel.Bioorg Med Chem Lett. 2023 Apr 1;85:129233. doi: 10.1016/j.bmcl.2023.129233. Epub 2023 Mar 9. Bioorg Med Chem Lett. 2023. PMID: 36905968 Free PMC article.
-
Proteasome substrate receptors and their therapeutic potential.Trends Biochem Sci. 2022 Nov;47(11):950-964. doi: 10.1016/j.tibs.2022.06.006. Epub 2022 Jul 9. Trends Biochem Sci. 2022. PMID: 35817651 Free PMC article. Review.
-
Chemical approaches to targeted protein degradation through modulation of the ubiquitin-proteasome pathway.Biochem J. 2017 Mar 15;474(7):1127-1147. doi: 10.1042/BCJ20160762. Biochem J. 2017. PMID: 28298557 Free PMC article. Review.
-
Measurement of the Multiple Activities of 26S Proteasomes.Methods Mol Biol. 2018;1844:289-308. doi: 10.1007/978-1-4939-8706-1_19. Methods Mol Biol. 2018. PMID: 30242717 Free PMC article.
-
Small-molecule control of intracellular protein levels through modulation of the ubiquitin proteasome system.Angew Chem Int Ed Engl. 2014 Feb 24;53(9):2312-30. doi: 10.1002/anie.201307761. Epub 2014 Jan 23. Angew Chem Int Ed Engl. 2014. PMID: 24459094 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

