PKR Mediates the Mitochondrial Unfolded Protein Response through Double-Stranded RNA Accumulation under Mitochondrial Stress
- PMID: 39062980
- PMCID: PMC11276775
- DOI: 10.3390/ijms25147738
PKR Mediates the Mitochondrial Unfolded Protein Response through Double-Stranded RNA Accumulation under Mitochondrial Stress
Abstract
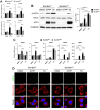
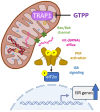
Mitochondrial stress, resulting from dysfunction and proteostasis disturbances, triggers the mitochondrial unfolded protein response (UPRMT), which activates gene encoding chaperones and proteases to restore mitochondrial function. Although ATFS-1 mediates mitochondrial stress UPRMT induction in C. elegans, the mechanisms relaying mitochondrial stress signals to the nucleus in mammals remain poorly defined. Here, we explored the role of protein kinase R (PKR), an eIF2α kinase activated by double-stranded RNAs (dsRNAs), in mitochondrial stress signaling. We found that UPRMT does not occur in cells lacking PKR, indicating its crucial role in this process. Mechanistically, we observed that dsRNAs accumulate within mitochondria under stress conditions, along with unprocessed mitochondrial transcripts. Furthermore, we demonstrated that accumulated mitochondrial dsRNAs in mouse embryonic fibroblasts (MEFs) deficient in the Bax/Bak channels are not released into the cytosol and do not induce the UPRMT upon mitochondrial stress, suggesting a potential role of the Bax/Bak channels in mediating the mitochondrial stress response. These discoveries enhance our understanding of how cells maintain mitochondrial integrity, respond to mitochondrial dysfunction, and communicate stress signals to the nucleus through retrograde signaling. This knowledge provides valuable insights into prospective therapeutic targets for diseases associated with mitochondrial stress.
Keywords: PKR; UPRMT; integrated stress response; mitochondrial dsRNAs; mitochondrial stress.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
RETRACTED: Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha.Science. 2006 Apr 28;312(5773):572-6. doi: 10.1126/science.1123480. Science. 2006. Retraction in: Science. 2024 Apr 19;384(6693):280. doi: 10.1126/science.adp1104. PMID: 16645094 Retracted.
-
Mitochondria Retrograde Signaling and the UPR mt: Where Are We in Mammals?Int J Mol Sci. 2015 Aug 6;16(8):18224-51. doi: 10.3390/ijms160818224. Int J Mol Sci. 2015. PMID: 26258774 Free PMC article. Review.
-
Bnip3 mediates mitochondrial dysfunction and cell death through Bax and Bak.Biochem J. 2007 Aug 1;405(3):407-15. doi: 10.1042/BJ20070319. Biochem J. 2007. PMID: 17447897 Free PMC article.
-
Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation.Gut. 2012 Sep;61(9):1269-1278. doi: 10.1136/gutjnl-2011-300767. Epub 2011 Oct 13. Gut. 2012. PMID: 21997551 Free PMC article.
-
Folding the Mitochondrial UPR into the Integrated Stress Response.Trends Cell Biol. 2020 Jun;30(6):428-439. doi: 10.1016/j.tcb.2020.03.001. Epub 2020 Apr 2. Trends Cell Biol. 2020. PMID: 32413314 Free PMC article. Review.
References
-
- King N. Amino Acids and the Mitochondria. In: Schaffer S., Suleiman M., editors. Advances in Biochemistry in Health and Disease, Mitochondria. Volume 2 Springer; New York, NY, USA: 2007.
-
- Nowinski S.M., Solmonson A., Rusin S.F., Maschek J.A., Bensard C.L., Fogarty S., Jeong M.Y., Lettlova S., Berg J.A., Morgan J.T., et al. Mitochondrial fatty acid synthesis coordinates oxidative metabolism in mammalian mitochondria. Elife. 2020;9:e58041. doi: 10.7554/eLife.58041. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

