Transcriptomic and Epigenomic Responses to Cortisol-Mediated Stress in Rainbow Trout (Oncorhynchus mykiss) Skeletal Muscle
- PMID: 39062828
- PMCID: PMC11276852
- DOI: 10.3390/ijms25147586
Transcriptomic and Epigenomic Responses to Cortisol-Mediated Stress in Rainbow Trout (Oncorhynchus mykiss) Skeletal Muscle
Abstract
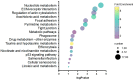
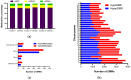
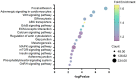
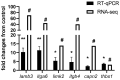
The production and release of cortisol during stress responses are key regulators of growth in teleosts. Understanding the molecular responses to cortisol is crucial for the sustainable farming of rainbow trout (Oncorhynchus mykiss) and other salmonid species. While several studies have explored the genomic and non-genomic impacts of cortisol on fish growth and skeletal muscle development, the long-term effects driven by epigenetic mechanisms, such as cortisol-induced DNA methylation, remain unexplored. In this study, we analyzed the transcriptome and genome-wide DNA methylation in the skeletal muscle of rainbow trout seven days after cortisol administration. We identified 550 differentially expressed genes (DEGs) by RNA-seq and 9059 differentially methylated genes (DMGs) via whole-genome bisulfite sequencing (WGBS) analysis. KEGG enrichment analysis showed that cortisol modulates the differential expression of genes associated with nucleotide metabolism, ECM-receptor interaction, and the regulation of actin cytoskeleton pathways. Similarly, cortisol induced the differential methylation of genes associated with focal adhesion, adrenergic signaling in cardiomyocytes, and Wnt signaling. Through integrative analyses, we determined that 126 genes showed a negative correlation between up-regulated expression and down-regulated methylation. KEGG enrichment analysis of these genes indicated participation in ECM-receptor interaction, regulation of actin cytoskeleton, and focal adhesion. Using RT-qPCR, we confirmed the differential expression of lamb3, itga6, limk2, itgb4, capn2, and thbs1. This study revealed for the first time the molecular responses of skeletal muscle to cortisol at the transcriptomic and whole-genome DNA methylation levels in rainbow trout.
Keywords: Oncorhynchus mykiss; RNA-seq; WGBS; cortisol; skeletal muscle.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
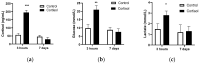
Similar articles
-
Early transcriptomic responses associated with the membrane-initiated action of cortisol in the skeletal muscle of rainbow trout (Oncorhynchus mykiss).Physiol Genomics. 2019 Nov 1;51(11):596-606. doi: 10.1152/physiolgenomics.00042.2019. Epub 2019 Oct 7. Physiol Genomics. 2019. PMID: 31588873
-
Regulation of the early expression of MAFbx/atrogin-1 and MuRF1 through membrane-initiated cortisol action in the skeletal muscle of rainbow trout.Comp Biochem Physiol B Biochem Mol Biol. 2021 Apr-May;253:110565. doi: 10.1016/j.cbpb.2021.110565. Epub 2021 Jan 23. Comp Biochem Physiol B Biochem Mol Biol. 2021. PMID: 33497801
-
Integrated Analyses of DNA Methylation and Gene Expression of Rainbow Trout Muscle under Variable Ploidy and Muscle Atrophy Conditions.Genes (Basel). 2022 Jun 26;13(7):1151. doi: 10.3390/genes13071151. Genes (Basel). 2022. PMID: 35885934 Free PMC article.
-
Membrane-initiated cortisol action modulates early pyruvate dehydrogenase kinase 2 (pdk2) expression in fish skeletal muscle.Comp Biochem Physiol A Mol Integr Physiol. 2019 Jul;233:24-29. doi: 10.1016/j.cbpa.2019.03.022. Epub 2019 Mar 28. Comp Biochem Physiol A Mol Integr Physiol. 2019. PMID: 30930204
-
Involvement of cortisol and sirtuin1 during the response to stress of hypothalamic circadian system and food intake-related peptides in rainbow trout, Oncorhynchus mykiss.Chronobiol Int. 2018 Aug;35(8):1122-1141. doi: 10.1080/07420528.2018.1461110. Epub 2018 May 8. Chronobiol Int. 2018. PMID: 29737878
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

