Rapid and Simple Morphological Assay for Determination of Susceptibility/Resistance to Combined Ciprofloxacin and Ampicillin, Independently, in Escherichia coli
- PMID: 39061357
- PMCID: PMC11273673
- DOI: 10.3390/antibiotics13070676
Rapid and Simple Morphological Assay for Determination of Susceptibility/Resistance to Combined Ciprofloxacin and Ampicillin, Independently, in Escherichia coli
Abstract
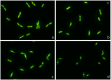
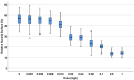
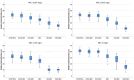
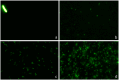
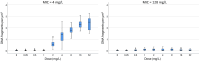
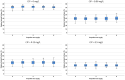
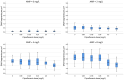
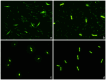
Current antibiograms cannot discern the particular effect of a specific antibiotic when the bacteria are incubated with a mixture of antibiotics. To prove that this task is achievable, Escherichia coli strains were treated with ciprofloxacin for 45 min, immobilized on a slide and stained with SYBR Gold. In susceptible strains, the nucleoid relative surface started to decrease near the MIC, being progressively condensed as the dose increased. The shrinkage level correlated with the DNA fragmentation degree. Ciprofloxacin-resistant bacilli showed no change. Additionally, E. coli strains were incubated with ampicillin for 45 min and processed similarly. The ampicillin-susceptible strain revealed intercellular DNA fragments that increased with dose, unlike the resistant strain. Co-incubation with both antibiotics revealed that ampicillin did not modify the nucleoid condensation effect of ciprofloxacin, whereas the quinolone partially decreased the background of DNA fragments induced by ampicillin. Sixty clinical isolates, with different combinations of susceptibility-resistance to each antibiotic, were co-incubated with the EUCAST breakpoints of susceptibility of ciprofloxacin and ampicillin. The morphological assay correctly categorized all the strains for each antibiotic in 60 min, demonstrating the feasible independent evaluation of a mixture of quinolone and beta-lactam. The rapid phenotypic assay may shorten the incubation times and necessary microbial mass currently required for evaluation.
Keywords: DNA fragments; antibiotic combination; bacterial nucleoid; beta-lactam; quinolone; rapid antibiogram.
Conflict of interest statement
J.G. and J.L.F. are advisers of Halotech DNA SL. All other authors declare no conflicts of interest.
Figures
Similar articles
-
A rapid in situ procedure for determination of bacterial susceptibility or resistance to antibiotics that inhibit peptidoglycan biosynthesis.BMC Microbiol. 2011 Aug 25;11:191. doi: 10.1186/1471-2180-11-191. BMC Microbiol. 2011. PMID: 21867549 Free PMC article.
-
Community-Acquired Uropathogenic Escherichia coli, Antimicrobial Susceptibility, and Extended-Spectrum Beta-Lactamase Detection.MEDICC Rev. 2022 May 16;24(2):20-25. doi: 10.37757/mr2022.v24.n2.2. MEDICC Rev. 2022. PMID: 35648059
-
Characterization of ciprofloxacin resistant Escherichia coli isolates among men undergoing evaluation for transrectal ultrasound guided prostate biopsy.J Urol. 2013 Dec;190(6):2026-32. doi: 10.1016/j.juro.2013.05.059. Epub 2013 May 30. J Urol. 2013. PMID: 23727416
-
Evaluation of synergistic activity of antibiotic combinations in extensive drug-resistant Acinetobacter species using checkerboard assay.J Med Microbiol. 2023 Feb;72(2). doi: 10.1099/jmm.0.001639. J Med Microbiol. 2023. PMID: 36762530
-
Phenotypic characteristics of coagulase-negative staphylococci: typing and antibiotic susceptibility.APMIS Suppl. 1999;91:1-42. APMIS Suppl. 1999. PMID: 10230367 Review.
References
-
- Cassini A., Högberg L.D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G.S., Colomb-Cotinat M., Kretzschmar M.E., Devleesschauwer B., Cecchini M., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

