Anticancer Effect of PtIIPHEN SS, PtII5ME SS, PtII56ME SS and Their Platinum(IV)-Dihydroxy Derivatives against Triple-Negative Breast Cancer and Cisplatin-Resistant Colorectal Cancer
- PMID: 39061185
- PMCID: PMC11274883
- DOI: 10.3390/cancers16142544
Anticancer Effect of PtIIPHEN SS, PtII5ME SS, PtII56ME SS and Their Platinum(IV)-Dihydroxy Derivatives against Triple-Negative Breast Cancer and Cisplatin-Resistant Colorectal Cancer
Abstract
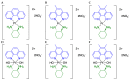
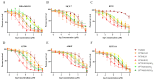
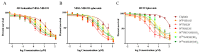
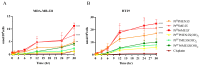
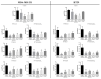
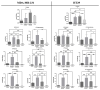
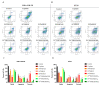
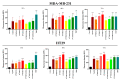
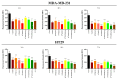
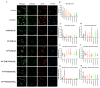
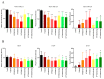
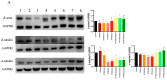
Development of resistance to cisplatin, oxaliplatin and carboplatin remains a challenge for their use as chemotherapies, particularly in breast and colorectal cancer. Here, we compare the anticancer effect of novel complexes [Pt(1,10-phenanthroline)(1S,2S-diaminocyclohexane)](NO3)2 (PtIIPHENSS), [Pt(5-methyl-1,10-phenanthroline)(1S,2S-diaminocyclohexane)](NO3)2 (PtII5MESS) and [Pt(5,6-dimethyl-1,10-phenanthroline)(1S,2S-diaminocyclohexane)](NO3)2 (PtII56MESS) and their platinum(IV)-dihydroxy derivatives with cisplatin. Complexes are greater than 11-fold more potent than cisplatin in both 2D and 3D cell line cultures with increased selectivity for cancer cells over genetically stable cells. ICP-MS studies showed cellular uptake occurred through an active transport mechanism with considerably altered platinum concentrations found in the cytoskeleton across all complexes after 24 h. Significant reactive oxygen species generation was observed, with reduced mitochondrial membrane potential at 72 h of treatment. Late apoptosis/necrosis was shown by Annexin V-FITC/PI flow cytometry assay, accompanied by increased sub-G0/G1 cells compared with untreated cells. An increase in S and G2+M cells was seen with all complexes. Treatment resulted in significant changes in actin and tubulin staining. Intrinsic and extrinsic apoptosis markers, MAPK/ERK and PI3K/AKT activation markers, together with autophagy markers showed significant activation of these pathways by Western blot. The proteomic profile investigated post-72 h of treatment identified 1597 MDA-MB-231 and 1859 HT29 proteins quantified by mass spectroscopy, with several differentially expressed proteins relative to no treatment. GO enrichment analysis revealed a statistically significant enrichment of RNA/DNA-associated proteins in both the cell lines and specific additional processes for individual drugs. This study shows that these novel agents function as multi-mechanistic chemotherapeutics, offering promising anticancer potential, and thereby supporting further research into their application as cancer therapeutics.
Keywords: breast cancer; chemotherapy; cisplatin resistant; colorectal cancer; cytotoxic; mechanism; platinum(II); platinum(IV); proteomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures













Similar articles
-
Chemotherapeutic Potential of Chlorambucil-Platinum(IV) Prodrugs against Cisplatin-Resistant Colorectal Cancer Cells.Int J Mol Sci. 2024 Jul 28;25(15):8252. doi: 10.3390/ijms25158252. Int J Mol Sci. 2024. PMID: 39125821 Free PMC article.
-
Potent Chlorambucil-Platinum(IV) Prodrugs.Int J Mol Sci. 2022 Sep 9;23(18):10471. doi: 10.3390/ijms231810471. Int J Mol Sci. 2022. PMID: 36142383 Free PMC article.
-
The in vitro renal cell toxicity of some unconventional anticancer phenanthroline-based platinum(II) complexes.J Inorg Biochem. 2018 Feb;179:97-106. doi: 10.1016/j.jinorgbio.2017.11.021. Epub 2017 Dec 1. J Inorg Biochem. 2018. PMID: 29197671
-
Potent Platinum(IV) Prodrugs That Incorporate a Biotin Moiety to Selectively Target Cancer Cells.Pharmaceutics. 2022 Dec 12;14(12):2780. doi: 10.3390/pharmaceutics14122780. Pharmaceutics. 2022. PMID: 36559273 Free PMC article.
-
Advancements in the Use of Platinum Complexes as Anticancer Agents.Anticancer Agents Med Chem. 2022;22(5):821-835. doi: 10.2174/1871520621666210805150705. Anticancer Agents Med Chem. 2022. PMID: 34353272 Review.
Cited by
-
Chemotherapeutic Potential of Chlorambucil-Platinum(IV) Prodrugs against Cisplatin-Resistant Colorectal Cancer Cells.Int J Mol Sci. 2024 Jul 28;25(15):8252. doi: 10.3390/ijms25158252. Int J Mol Sci. 2024. PMID: 39125821 Free PMC article.
References
-
- World Health Organization Cancer. [(accessed on 7 March 2024)]. Available online: https://www.who.int/health-topics/cancer#tab=tab_1.
-
- Roser M., Ritchie H. Cancer. Our World in Data. 2015. [(accessed on 7 March 2024)]. Available online: https://ourworldindata.org/cancer.
-
- Wiltshaw E. Cisplatin in the treatment of cancer-the first metal anti-tumour drug. Platin. Met. Rev. 1979;23:90–98. doi: 10.1595/003214079X2339098. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

