Structure-based humanization of a therapeutic antibody for multiple myeloma
- PMID: 39052065
- PMCID: PMC11358308
- DOI: 10.1007/s00109-024-02470-4
Structure-based humanization of a therapeutic antibody for multiple myeloma
Abstract
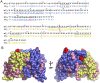
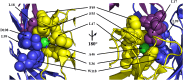
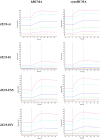
The optimal efficacy of xenogeneically generated proteins intended for application in humans requires that their own antigenicity be minimized. This necessary adaptation of antibodies to a humanized version poses challenges since modifications even distant from the binding sites can greatly influence antigen recognition and this is the primary feature that must be maintained during all modifications. Current strategies often rely on grafting and/or randomization/selection to arrive at a humanized variant retaining the binding properties of the original molecule. However, in terms of speed and efficiency, rationally directed approaches can be superior, provided the requisite structural information is available. We present here a humanization procedure based on the high-resolution X-ray structure of a chimaeric IgG against a marker for multiple myeloma. Based on in silico modelling of humanizing amino acid substitutions identified from sequence alignments, we devised a straightforward cloning procedure to rapidly evaluate the proposed sequence changes. Careful inspection of the structure allowed the identification of a potentially problematic amino acid change that indeed disrupted antigen binding. Subsequent optimization of the antigen binding loop sequences resulted in substantial recovery of binding affinity lost in the completely humanized antibody. X-ray structures of the humanized and optimized variants demonstrate that the antigen binding mode is preserved, with surprisingly few direct contacts to antibody atoms. These results underline the importance of structural information for the efficient optimization of protein therapeutics. KEY MESSAGES: Structure-based humanization of an IgG against BCMA, a marker for Multiple Myeloma. Identification of problematic mutations and unexpected modification sites. Structures of the modified IgG-antigen complexes verified predictions. Provision of humanized high-affinity IgGs against BCMA for therapeutic applications.
Keywords: Humanization; Monoclonal antibody; Multiple myeloma; X-ray structure.
© 2024. The Author(s).
Conflict of interest statement
SFM and OD are inventors on multiple pending and granted patents derived from the international patent applications PCT/EP2013/072857, PCT/EP2015/059562, and PCT/EP2017/063862 concerning J22.9-xi and variants.
Figures
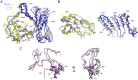
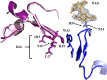
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Far Posterior Approach for Rib Fracture Fixation: Surgical Technique and Tips.JBJS Essent Surg Tech. 2024 Dec 6;14(4):e23.00094. doi: 10.2106/JBJS.ST.23.00094. eCollection 2024 Oct-Dec. JBJS Essent Surg Tech. 2024. PMID: 39650795 Free PMC article.
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7 PMID: 38260445 Free PMC article. Updated. Preprint.
-
Australia in 2030: what is our path to health for all?Med J Aust. 2021 May;214 Suppl 8:S5-S40. doi: 10.5694/mja2.51020. Med J Aust. 2021. PMID: 33934362
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
References
-
- Lo K-M, Leger O, Hock B. Antibody engineering. Microbiol Spectrum 2(1):AID-0007–2012 (2014). 10.1128/microbiolspec.AID-0007-12 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

