Chlamydiae as symbionts of photosynthetic dinoflagellates
- PMID: 39046276
- PMCID: PMC11317633
- DOI: 10.1093/ismejo/wrae139
Chlamydiae as symbionts of photosynthetic dinoflagellates
Abstract
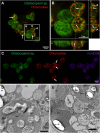
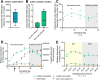
Chlamydiae are ubiquitous intracellular bacteria and infect a wide diversity of eukaryotes, including mammals. However, chlamydiae have never been reported to infect photosynthetic organisms. Here, we describe a novel chlamydial genus and species, Candidatus Algichlamydia australiensis, capable of infecting the photosynthetic dinoflagellate Cladocopium sp. (originally isolated from a scleractinian coral). Algichlamydia australiensis was confirmed to be intracellular by fluorescence in situ hybridization and confocal laser scanning microscopy and temporally stable at the population level by monitoring its relative abundance across four weeks of host growth. Using a combination of short- and long-read sequencing, we recovered a high-quality (completeness 91.73% and contamination 0.27%) metagenome-assembled genome of A. australiensis. Phylogenetic analyses show that this chlamydial taxon represents a new genus and species within the Simkaniaceae family. Algichlamydia australiensis possesses all the hallmark genes for chlamydiae-host interactions, including a complete type III secretion system. In addition, a type IV secretion system is encoded on a plasmid and has previously been observed for only three other chlamydial species. Twenty orthologous groups of genes are unique to A. australiensis, one of which is structurally similar to a protein known from Cyanobacteria and Archaeplastida involved in thylakoid biogenesis and maintenance, hinting at potential chlamydiae interactions with the chloroplasts of Cladocopium cells. Our study shows that chlamydiae infect dinoflagellate symbionts of cnidarians, the first photosynthetic organism reported to harbor chlamydiae, thereby expanding the breadth of chlamydial hosts and providing a new contribution to the discussion around the role of chlamydiae in the establishment of the primary plastid.
Keywords: Symbiodiniaceae; alga; chlamydiae; coral; genomics.
© The Author(s) 2024. Published by Oxford University Press on behalf of the International Society for Microbial Ecology.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Unity in variety--the pan-genome of the Chlamydiae.Mol Biol Evol. 2011 Dec;28(12):3253-70. doi: 10.1093/molbev/msr161. Epub 2011 Jun 20. Mol Biol Evol. 2011. PMID: 21690563 Free PMC article.
-
Chlamydial seasonal dynamics and isolation of 'Candidatus Neptunochlamydia vexilliferae' from a Tyrrhenian coastal lake.Environ Microbiol. 2016 Sep;18(8):2405-17. doi: 10.1111/1462-2920.13111. Epub 2015 Dec 21. Environ Microbiol. 2016. PMID: 26530333
-
Mutualistic Interactions between Dinoflagellates and Pigmented Bacteria Mitigate Environmental Stress.Microbiol Spectr. 2023 Feb 14;11(1):e0246422. doi: 10.1128/spectrum.02464-22. Epub 2023 Jan 18. Microbiol Spectr. 2023. PMID: 36651852 Free PMC article.
-
Chlamydiae as symbionts in eukaryotes.Annu Rev Microbiol. 2008;62:113-31. doi: 10.1146/annurev.micro.62.081307.162818. Annu Rev Microbiol. 2008. PMID: 18473699 Review.
-
Emerging chlamydial infections.Crit Rev Microbiol. 2004;30(2):75-106. doi: 10.1080/10408410490435106. Crit Rev Microbiol. 2004. PMID: 15239381 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

