Suppression of interferon α and γ response by Huwe1-mediated Miz1 degradation promotes SARS-CoV-2 replication
- PMID: 39034993
- PMCID: PMC11257858
- DOI: 10.3389/fimmu.2024.1388517
Suppression of interferon α and γ response by Huwe1-mediated Miz1 degradation promotes SARS-CoV-2 replication
Abstract
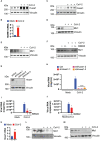
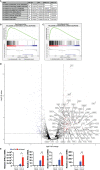
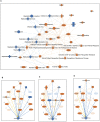
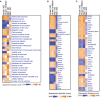
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has been demonstrated to limit the host interferon response; however, the underlying mechanism remains unclear. Here, we found that SARS-CoV-2 infection upregulated the E3 ubiquitin ligase Huwe1, which in turn facilitated the degradation of the transcription factor Miz1. The degradation of Miz1 hampered interferon alpha and gamma responses, consequently fostering viral replication and impeding viral clearance. Conversely, silencing or inhibiting Huwe1 enhanced the interferon responses, effectively curbing viral replication. Consistently, overexpressing Miz1 augmented the interferon responses and limited viral replication, whereas silencing Miz1 had the opposite effect. Targeting Huwe1 or overexpressing Miz1 elicited transcriptomic alterations characterized by enriched functions associated with bolstered antiviral response and diminished virus replication. Further study revealed Miz1 exerted epigenetic control over the transcription of specific interferon signaling molecules, which acted as common upstream regulators responsible for the observed transcriptomic changes following Huwe1 or Miz1 targeting. These findings underscore the critical role of the Huwe1-Miz1 axis in governing the host antiviral response, with its dysregulation contributing to the impaired interferon response observed during COVID-19.
Keywords: HUWE1; Miz1; SARS-CoV-2; interferon alpha and gamma responses; transcriptional regulation.
Copyright © 2024 Arunagiri, Cooper, Dong, Class, Biswas, Vahora, Deshpande, Gopani, Hu, Richner, Rong and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




Similar articles
-
Miz1 represses type I interferon production and limits viral clearance during influenza A virus infection.Sci Signal. 2024 Apr 9;17(831):eadg7867. doi: 10.1126/scisignal.adg7867. Epub 2024 Apr 9. Sci Signal. 2024. PMID: 38593156 Free PMC article.
-
Multiple E3 ligases act as antiviral factors against SARS-CoV-2 via inducing the ubiquitination and degradation of ORF9b.J Virol. 2024 Jun 13;98(6):e0162423. doi: 10.1128/jvi.01624-23. Epub 2024 May 6. J Virol. 2024. PMID: 38709105 Free PMC article.
-
Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15.Genes Dev. 2013 May 15;27(10):1101-14. doi: 10.1101/gad.214577.113. Genes Dev. 2013. PMID: 23699408 Free PMC article.
-
Spatial and temporal roles of SARS-CoV PLpro -A snapshot.FASEB J. 2021 Jan;35(1):e21197. doi: 10.1096/fj.202002271. FASEB J. 2021. PMID: 33368679 Free PMC article. Review.
-
Transcriptional and Non-Transcriptional Activation, Posttranslational Modifications, and Antiviral Functions of Interferon Regulatory Factor 3 and Viral Antagonism by the SARS-Coronavirus.Viruses. 2021 Mar 29;13(4):575. doi: 10.3390/v13040575. Viruses. 2021. PMID: 33805458 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

