All (remains) in the family? Using healthy relatives to define Crohn's gut microbiome alterations
- PMID: 39019007
- PMCID: PMC11293313
- DOI: 10.1016/j.xcrm.2024.101651
All (remains) in the family? Using healthy relatives to define Crohn's gut microbiome alterations
Abstract
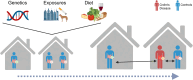
Gut microbial imbalance is noted in Crohn's disease (CD), but the specific bacteria associated with CD vary between studies. Chen et al.1 pair CD patients with their healthy first-degree relatives to mitigate some of the environmental and genetic effects.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
Similar articles
-
Altered Gut Microbiome Composition and Function Are Associated With Gut Barrier Dysfunction in Healthy Relatives of Patients With Crohn's Disease.Gastroenterology. 2022 Nov;163(5):1364-1376.e10. doi: 10.1053/j.gastro.2022.07.004. Epub 2022 Jul 16. Gastroenterology. 2022. PMID: 35850197
-
The Gut Microbial Profile of Preclinical Crohn's Disease Is Similar to That of Healthy Controls.Inflamm Bowel Dis. 2020 Oct 23;26(11):1682-1690. doi: 10.1093/ibd/izaa072. Inflamm Bowel Dis. 2020. PMID: 32339246
-
Gut Microbial Signatures Underline Complicated Crohn's Disease but Vary Between Cohorts; An In Silico Approach.Inflamm Bowel Dis. 2019 Jan 10;25(2):217-225. doi: 10.1093/ibd/izy328. Inflamm Bowel Dis. 2019. PMID: 30346536
-
The Microbiome in Crohn's Disease: Role in Pathogenesis and Role of Microbiome Replacement Therapies.Gastroenterol Clin North Am. 2017 Sep;46(3):481-492. doi: 10.1016/j.gtc.2017.05.004. Epub 2017 Jul 19. Gastroenterol Clin North Am. 2017. PMID: 28838410 Review.
-
Dysbiotic gut microbiome: A key element of Crohn's disease.Comp Immunol Microbiol Infect Dis. 2015 Dec;43:36-49. doi: 10.1016/j.cimid.2015.10.005. Epub 2015 Oct 25. Comp Immunol Microbiol Infect Dis. 2015. PMID: 26616659 Review.
References
-
- Chen W., Li Y., Wang W., Gao S., Hu J., Xiang B., Wu D., Jiao N., Xu T., Zhi M., et al. Enhanced microbiota profiling in patients with quiescent Crohn’s disease through comparison with paired healthy first-degree relatives. Cell Rep. Med. 2024;5:101624. doi: 10.1016/j.xcrm.2024.101624. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

