Single-cell level LasR-mediated quorum sensing response of Pseudomonas aeruginosa to pulses of signal molecules
- PMID: 39003361
- PMCID: PMC11246452
- DOI: 10.1038/s41598-024-66706-6
Single-cell level LasR-mediated quorum sensing response of Pseudomonas aeruginosa to pulses of signal molecules
Abstract
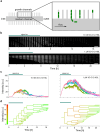
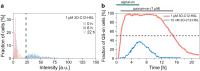
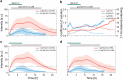
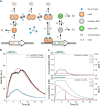
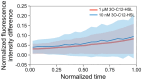
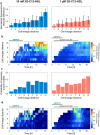
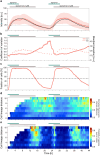
Quorum sensing (QS) is a communication form between bacteria via small signal molecules that enables global gene regulation as a function of cell density. We applied a microfluidic mother machine to study the kinetics of the QS response of Pseudomonas aeruginosa bacteria to additions and withdrawals of signal molecules. We traced the fast buildup and the subsequent considerably slower decay of a population-level and single-cell-level QS response. We applied a mathematical model to explain the results quantitatively. We found significant heterogeneity in QS on the single-cell level, which may result from variations in quorum-controlled gene expression and protein degradation. Heterogeneity correlates with cell lineage history, too. We used single-cell data to define and quantitatively characterize the population-level quorum state. We found that the population-level QS response is well-defined. The buildup of the quorum is fast upon signal molecule addition. At the same time, its decay is much slower following signal withdrawal, and the quorum may be maintained for several hours in the absence of the signal. Furthermore, the quorum sensing response of the population was largely repeatable in subsequent pulses of signal molecules.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
LasR Variant Cystic Fibrosis Isolates Reveal an Adaptable Quorum-Sensing Hierarchy in Pseudomonas aeruginosa.mBio. 2016 Oct 4;7(5):e01513-16. doi: 10.1128/mBio.01513-16. mBio. 2016. PMID: 27703072 Free PMC article.
-
Links between Anr and Quorum Sensing in Pseudomonas aeruginosa Biofilms.J Bacteriol. 2015 Sep;197(17):2810-20. doi: 10.1128/JB.00182-15. Epub 2015 Jun 15. J Bacteriol. 2015. PMID: 26078448 Free PMC article.
-
Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database.Antimicrob Agents Chemother. 2013 Nov;57(11):5629-41. doi: 10.1128/AAC.00955-13. Epub 2013 Sep 3. Antimicrob Agents Chemother. 2013. PMID: 24002091 Free PMC article.
-
An evolving perspective on the Pseudomonas aeruginosa orphan quorum sensing regulator QscR.Front Cell Infect Microbiol. 2014 Oct 28;4:152. doi: 10.3389/fcimb.2014.00152. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 25389523 Free PMC article. Review.
-
Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules.Curr Opin Microbiol. 2009 Apr;12(2):182-91. doi: 10.1016/j.mib.2009.01.005. Epub 2009 Feb 25. Curr Opin Microbiol. 2009. PMID: 19249239 Review.
References
MeSH terms
Substances
Grants and funding
- K116516/Nemzeti Kutatási, Fejlesztési és Innovációs Hivatal (NKFI Office)
- FK138520/Nemzeti Kutatási, Fejlesztési és Innovációs Hivatal (NKFI Office)
- PD124889/Nemzeti Kutatási, Fejlesztési és Innovációs Hivatal (NKFI Office)
- BO/00290/21/11/Magyar Tudományos Akadémia (Hungarian Academy of Sciences)
- BO/00463/18/8/Magyar Tudományos Akadémia (Hungarian Academy of Sciences)
LinkOut - more resources
Full Text Sources

