Arctigenin from Forsythia viridissima Fruit Inhibits the Replication of Human Coronavirus
- PMID: 39000469
- PMCID: PMC11242317
- DOI: 10.3390/ijms25137363
Arctigenin from Forsythia viridissima Fruit Inhibits the Replication of Human Coronavirus
Abstract
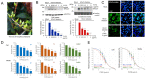
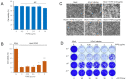
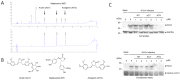
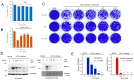
Coronavirus can cause various diseases, from mild symptoms to the recent severe COVID-19. The coronavirus RNA genome is frequently mutated due to its RNA nature, resulting in many pathogenic and drug-resistant variants. Therefore, many medicines should be prepared to respond to the various coronavirus variants. In this report, we demonstrated that Forsythia viridissima fruit ethanol extract (FVFE) effectively reduces coronavirus replication. We attempted to identify the active compounds and found that actigenin from FVFE effectively reduces human coronavirus replication. Arctigenin treatment can reduce coronavirus protein expression and coronavirus-induced cytotoxicity. These results collectively suggest that arctigenin is a potent natural compound that prevents coronavirus replication.
Keywords: antiviral; arctigenin; forsythia viridissima; human coronavirus; natural compound.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Phillyrin (KD-1) exerts anti-viral and anti-inflammatory activities against novel coronavirus (SARS-CoV-2) and human coronavirus 229E (HCoV-229E) by suppressing the nuclear factor kappa B (NF-κB) signaling pathway.Phytomedicine. 2020 Nov;78:153296. doi: 10.1016/j.phymed.2020.153296. Epub 2020 Aug 1. Phytomedicine. 2020. PMID: 32890913 Free PMC article.
-
Arctigenin from Arctium lappa L. inhibits chikungunya virus by affecting its entry and replication.Phytomedicine. 2024 Jun;128:155491. doi: 10.1016/j.phymed.2024.155491. Epub 2024 Feb 27. Phytomedicine. 2024. PMID: 38489894
-
The Petasites hybridus CO2 Extract (Ze 339) Blocks SARS-CoV-2 Replication In Vitro.Viruses. 2022 Jan 7;14(1):106. doi: 10.3390/v14010106. Viruses. 2022. PMID: 35062310 Free PMC article.
-
Antiasthmatic action of dibenzylbutyrolactone lignans from fruits of Forsythia viridissima on asthmatic responses to ovalbumin challenge in conscious guinea-pigs.Phytother Res. 2011 Mar;25(3):387-95. doi: 10.1002/ptr.3273. Epub 2010 Aug 23. Phytother Res. 2011. PMID: 20734328
-
A Brief Overview of Potential Treatments for Viral Diseases Using Natural Plant Compounds: The Case of SARS-Cov.Molecules. 2021 Jun 24;26(13):3868. doi: 10.3390/molecules26133868. Molecules. 2021. PMID: 34202844 Free PMC article. Review.
Cited by
-
Natural products and derivatives as Japanese encephalitis virus antivirals.Pathog Dis. 2024 Feb 7;82:ftae022. doi: 10.1093/femspd/ftae022. Pathog Dis. 2024. PMID: 39317665 Free PMC article. Review.
References
-
- Harrison C.M., Doster J.M., Landwehr E.H., Kumar N.P., White E.J., Beachboard D.C., Stobart C.C. Evaluating the Virology and Evolution of Seasonal Human Coronaviruses Associated with the Common Cold in the COVID-19 Era. Microorganisms. 2023;11:445. doi: 10.3390/microorganisms11020445. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

