Proteomic Changes Induced by the Immunosuppressant Everolimus in Human Podocytes
- PMID: 39000447
- PMCID: PMC11242170
- DOI: 10.3390/ijms25137336
Proteomic Changes Induced by the Immunosuppressant Everolimus in Human Podocytes
Abstract
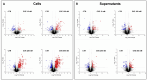
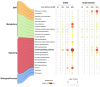
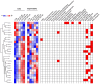
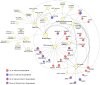
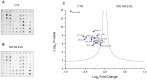
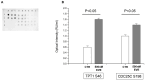
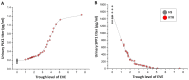
mTOR inhibitors (mTOR-Is) may induce proteinuria in kidney transplant recipients through podocyte damage. However, the mechanism has only been partially defined. Total cell lysates and supernatants of immortalized human podocytes treated with different doses of everolimus (EVE) (10, 100, 200, and 500 nM) for 24 h were subjected to mass spectrometry-based proteomics. Support vector machine and partial least squares discriminant analysis were used for data analysis. The results were validated in urine samples from 28 kidney transplant recipients receiving EVE as part of their immunosuppressive therapy. We identified more than 7000 differentially expressed proteins involved in several pathways, including kinases, cell cycle regulation, epithelial-mesenchymal transition, and protein synthesis, according to gene ontology. Among these, after statistical analysis, 65 showed an expression level significantly and directly correlated with EVE dosage. Polo-Like Kinase 1 (PLK1) content was increased, whereas osteopontin (SPP1) content was reduced in podocytes and supernatants in a dose-dependent manner and significantly correlated with EVE dose (p < 0.0001, FDR < 5%). Similar results were obtained in the urine of kidney transplant patients. This study analyzed the impact of different doses of mTOR-Is on podocytes, helping to understand not only the biological basis of their therapeutic effects but also the possible mechanisms underlying proteinuria.
Keywords: everolimus; kidney transplantation; mTOR-inhibitors; podocytes; proteomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Cellular effects of everolimus and sirolimus on podocytes.PLoS One. 2013 Nov 15;8(11):e80340. doi: 10.1371/journal.pone.0080340. eCollection 2013. PLoS One. 2013. PMID: 24260371 Free PMC article.
-
Everolimus-induced epithelial to mesenchymal transition (EMT) in bronchial/pulmonary cells: when the dosage does matter in transplantation.J Nephrol. 2016 Dec;29(6):881-891. doi: 10.1007/s40620-016-0295-4. Epub 2016 Mar 29. J Nephrol. 2016. PMID: 27026415
-
Comparative Proteomic Analysis of Rapamycin Versus Cyclosporine Combination Treatment in Mouse Podocytes.Transplant Proc. 2016 May;48(4):1297-301. doi: 10.1016/j.transproceed.2016.01.022. Transplant Proc. 2016. PMID: 27320608
-
Calcineurin inhibitor-free immunosuppression in pediatric renal transplantation: a viable option?Paediatr Drugs. 2011 Feb 1;13(1):49-69. doi: 10.2165/11538530-000000000-00000. Paediatr Drugs. 2011. PMID: 21162600 Review.
-
Direct Effects of Immunomodulatory Agents on Podocytes in Immune-Mediated Glomerular Diseases.Contrib Nephrol. 2018;195:131-142. doi: 10.1159/000486943. Epub 2018 May 7. Contrib Nephrol. 2018. PMID: 29734158 Review.
References
-
- Schena F.P., Pascoe M.D., Alberu J., del Carmen Rial M., Oberbauer R., Brennan D.C., Campistol J.M., Racusen L., Polinsky M.S., Goldberg-Alberts R., et al. Sirolimus CONVERT Trial Study Group. Conversion from calcineurin inhibitors to sirolimus maintenance therapy in renal allograft recipients: 24-month efficacy and safety results from the CONVERT trial. Transplantation. 2009;87:233–242. doi: 10.1097/TP.0b013e3181927a41. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

