IRF8-mutant B cell lymphoma evades immunity through a CD74-dependent deregulation of antigen processing and presentation in MHCII complexes
- PMID: 38996030
- PMCID: PMC11244530
- DOI: 10.1126/sciadv.adk2091
IRF8-mutant B cell lymphoma evades immunity through a CD74-dependent deregulation of antigen processing and presentation in MHCII complexes
Abstract
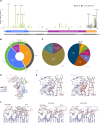
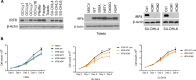
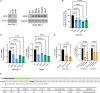
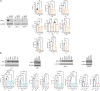
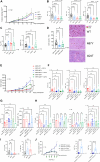
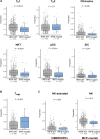
The mechanism by which interferon regulatory factor 8 (IRF8) mutation contributes to lymphomagenesis is unknown. We modeled IRF8 variants in B cell lymphomas and found that they affected the expression of regulators of antigen presentation. Expression of IRF8 mutants in murine B cell lymphomas suppressed CD4, but not CD8, activation elicited by antigen presentation and downmodulated CD74 and human leukocyte antigen (HLA) DM, intracellular regulators of antigen peptide processing/loading in the major histocompatibility complex (MHC) II. Concordantly, mutant IRF8 bound less efficiently to the promoters of these genes. Mice harboring IRF8 mutant lymphomas displayed higher tumor burden and remodeling of the tumor microenvironment, typified by depletion of CD4, CD8, and natural killer cells, increase in regulatory T cells and T follicular helper cells. Deconvolution of bulk RNA sequencing data from IRF8-mutant human diffuse large B cell lymphoma (DLBCL) recapitulated part of the immune remodeling detected in mice. We concluded that IRF8 mutations contribute to DLBCL biology by facilitating immune escape.
Figures

Update of
-
IRF8-mutant B cell lymphoma evades immunity through a CD74-dependent deregulation of antigen processing and presentation in MHC CII complexes.bioRxiv [Preprint]. 2023 Oct 15:2023.10.14.560755. doi: 10.1101/2023.10.14.560755. bioRxiv. 2023. Update in: Sci Adv. 2024 Jul 12;10(28):eadk2091. doi: 10.1126/sciadv.adk2091 PMID: 37873241 Free PMC article. Updated. Preprint.
Similar articles
-
IRF8-mutant B cell lymphoma evades immunity through a CD74-dependent deregulation of antigen processing and presentation in MHC CII complexes.bioRxiv [Preprint]. 2023 Oct 15:2023.10.14.560755. doi: 10.1101/2023.10.14.560755. bioRxiv. 2023. Update in: Sci Adv. 2024 Jul 12;10(28):eadk2091. doi: 10.1126/sciadv.adk2091 PMID: 37873241 Free PMC article. Updated. Preprint.
-
CD74 promotes the formation of an immunosuppressive tumor microenvironment in triple-negative breast cancer in mice by inducing the expansion of tolerogenic dendritic cells and regulatory B cells.PLoS Biol. 2024 Nov 22;22(11):e3002905. doi: 10.1371/journal.pbio.3002905. eCollection 2024 Nov. PLoS Biol. 2024. PMID: 39576827 Free PMC article.
-
Interferon regulatory factor 8-driven reprogramming of the immune microenvironment enhances antitumor adaptive immunity and reduces immunosuppression in murine glioblastoma.Neuro Oncol. 2024 Dec 5;26(12):2272-2287. doi: 10.1093/neuonc/noae149. Neuro Oncol. 2024. PMID: 39115195
-
Antigen presentation plays positive roles in the regenerative response to cardiac injury in zebrafish.Nat Commun. 2024 Apr 29;15(1):3637. doi: 10.1038/s41467-024-47430-1. Nat Commun. 2024. PMID: 38684665 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
References
-
- Thorsson V., Gibbs D. L., Brown S. D., Wolf D., Bortone D. S., Yang T. H. O., Porta-Pardo E., Gao G. F., Plaisier C. L., Eddy J. A., Ziv E., Culhane A. C., Paull E. O., Sivakumar I. K. A., Gentles A. J., Malhotra R., Farshidfar F., Colaprico A., Parker J. S., Mose L. E., Vo N. S., Liu J., Liu Y., Rader J., Dhankani V., Reynolds S. M., Bowlby R., Califano A., Cherniack A. D., Anastassiou D., Bedognetti D., Mokrab Y., Newman A. M., Rao A., Chen K., Krasnitz A., Hu H., Malta T. M., Noushmehr H., Pedamallu C. S., Bullman S., Ojesina A. I., Lamb A., Zhou W., Shen H., Choueiri T. K., Weinstein J. N., Guinney J., Saltz J., Holt R. A., Rabkin C. S., Cancer Genome Atlas Research Network, Lazar A. J., Serody J. S., Demicco E. G., Disis M. L., Vincent B. G., Shmulevich I., The immune landscape of cancer. Immunity 48, 812–830.e14 (2018). - PMC - PubMed
-
- Wellenstein M. D., de Visser K. E., Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity 48, 399–416 (2018). - PubMed
-
- Sharma P., Goswami S., Raychaudhuri D., Siddiqui B. A., Singh P., Nagarajan A., Liu J., Subudhi S. K., Poon C., Gant K. L., Herbrich S. M., Anandhan S., Islam S., Amit M., Anandappa G., Allison J. P., Immune checkpoint therapy-current perspectives and future directions. Cell 186, 1652–1669 (2023). - PubMed
-
- Challa-Malladi M., Lieu Y. K., Califano O., Holmes A. B., Bhagat G., Murty V. V., Dominguez-Sola D., Pasqualucci L., Dalla-Favera R., Combined genetic inactivation of β2-microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell 20, 728–740 (2011). - PMC - PubMed
-
- Steidl C., Shah S. P., Woolcock B. W., Rui L., Kawahara M., Farinha P., Johnson N. A., Zhao Y., Telenius A., Neriah S. B., McPherson A., Meissner B., Okoye U. C., Diepstra A., van den Berg A., Sun M., Leung G., Jones S. J., Connors J. M., Huntsman D. G., Savage K. J., Rimsza L. M., Horsman D. E., Staudt L. M., Steidl U., Marra M. A., Gascoyne R. D., MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 471, 377–381 (2011). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

