Molecular characterization of EcCLP1, a new putative cathepsin L protease from Echinococcus canadensis
- PMID: 38995112
- PMCID: PMC11242924
- DOI: 10.1051/parasite/2024036
Molecular characterization of EcCLP1, a new putative cathepsin L protease from Echinococcus canadensis
Abstract
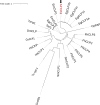
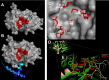
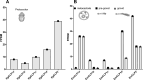
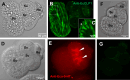
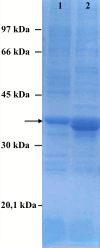
Echinococcus granulosus sensu lato is a platyhelminth parasite and the etiological cause of cystic echinococcosis (CE), a zoonotic and neglected disease that infects animals and humans worldwide. As a part of the biological arsenal of the parasite, cathepsin L proteases are a group of proteins that are believed to be essential for parasite penetration, immune evasion, and establishment in the tissues of the host. In this work, we have cloned and sequenced a new putative cathepsin L protease from Echinococcus canadensis (EcCLP1). The bioinformatic analysis suggests that EcCLP1 could be synthesized as a zymogen and activated after proteolytic cleavage. The multiple sequence alignment with other cathepsin proteases reveals important functional conserved features like a conserved active site, an N-linked glycosylation residue, a catalytic triad, an oxyanion hole, and three putative disulfide bonds. The phylogenetic analysis suggests that EcCLP1 could indeed be a cathepsin L cysteine protease from clade 1 as it grouped with cathepsins from other species in this clade. Modeling studies suggest that EcCLP1 has two domains forming a cleft where the active site is located and an occluding role for the propeptide. The transcriptomic analysis reveals different levels of cathepsin transcript expression along the different stages of the parasite life cycle. The whole-mount immunohistochemistry shows an interesting superficial punctate pattern of staining which suggests a secretory pattern of expression. The putative cathepsin L protease characterized here may represent an interesting tool for diagnostic purposes, vaccine design, or a new pharmacological target for antiparasitic intervention.
Title: Caractérisation moléculaire d’EcCLP1, une nouvelle protéase putative de type cathepsine L d’Echinococcus canadensis.
Abstract: Echinococcus granulosus sensu lato est un Plathelminthe parasite et la cause étiologique de l’échinococcose kystique (EK), une maladie zoonotique et négligée qui infecte les animaux et les humains dans le monde entier. En tant que partie de l’arsenal biologique du parasite, les protéases de type cathepsine L sont un groupe de protéines considérées comme essentielles à la pénétration du parasite, l’évasion immunitaire et son établissement dans les tissus de l’hôte. Dans ce travail, nous avons cloné et séquencé une nouvelle protéase putative de type cathepsine L d’Echinococcus canadensis (EcCLP1). L’analyse bioinformatique suggère qu’EcCLP1 pourrait être synthétisée sous forme de zymogène et activée après clivage protéolytique. L’alignement de séquences multiples avec d’autres protéases de type cathepsine révèle d’importantes caractéristiques fonctionnelles conservées telles qu’un site actif conservé, un résidu de glycosylation lié à N, une triade catalytique, un trou oxyanion et trois liaisons disulfure putatives. L’analyse phylogénétique suggère qu’EcCLP1 pourrait en effet être une protéase de type cathepsine L du clade 1 car elle se regroupe avec les cathepsines d’autres espèces de ce clade. Les études de modélisation suggèrent qu’EcCLP1 possède deux domaines formant une fente où se trouve le site actif et un rôle d’occlusion pour le propeptide. L’analyse transcriptomique révèle différents niveaux d’expression du transcrit de la cathepsine au cours des différentes étapes du cycle de vie du parasite. L’immunohistochimie de montages entiers montre un intéressant motif de coloration ponctuée superficielle qui suggère un modèle d’expression sécrétoire. La protéase putative de type cathepsine L caractérisée ici peut représenter un outil intéressant à des fins de diagnostic, de conception de vaccins ou une nouvelle cible pharmacologique pour une intervention antiparasitaire.
Keywords: Cathepsin L proteases; Echinococcus canadensis; Echinococcus granulosus sensu lato; Propeptide.
© A. Naidich et al., published by EDP Sciences, 2024.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Comparison between Echinococcus granulosus sensu stricto (G1) and E. canadensis (G6) mitochondrial genes (cox1 and nad1) and their related protein models using experimental and bioinformatics analysis.Comput Biol Chem. 2019 Apr;79:103-109. doi: 10.1016/j.compbiolchem.2019.02.002. Epub 2019 Feb 5. Comput Biol Chem. 2019. PMID: 30769268
-
The Echinococcus canadensis (G7) genome: a key knowledge of parasitic platyhelminth human diseases.BMC Genomics. 2017 Feb 27;18(1):204. doi: 10.1186/s12864-017-3574-0. BMC Genomics. 2017. PMID: 28241794 Free PMC article.
-
Genetic variation of Echinococcus spp. in yaks and sheep in the Tibet Autonomous Region of China based on mitochondrial DNA.Parasit Vectors. 2019 Dec 27;12(1):608. doi: 10.1186/s13071-019-3857-1. Parasit Vectors. 2019. PMID: 31881922 Free PMC article.
-
Echinococcus granulosus sensu lato genotypes infecting humans--review of current knowledge.Int J Parasitol. 2014 Jan;44(1):9-18. doi: 10.1016/j.ijpara.2013.08.008. Epub 2013 Nov 19. Int J Parasitol. 2014. PMID: 24269720 Review.
-
Cystic echinococcosis in South America: systematic review of species and genotypes of Echinococcus granulosus sensu lato in humans and natural domestic hosts.Trop Med Int Health. 2016 Feb;21(2):166-75. doi: 10.1111/tmi.12647. Epub 2015 Dec 28. Trop Med Int Health. 2016. PMID: 26610060 Review.
References
-
- Baig S, Damian RT, Molinari JL, Tato P, Morales-Montor J, Welch M, Talhouk J, Hashmeys R, White AC Jr. 2005. Purification and characterization of a metacestode cysteine proteinase from Taenia solium involved in the breakdown of human IgG. Parasitology, 131(Pt 3), 411–416. - PubMed
-
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research, 42(Web Server issue), W252–W258. - PMC - PubMed
-
- Camicia F, Paredes R, Chalar C, Galanti N, Kamenetzky L, Gutierrez A, Rosenzvit MC. 2008. Sequencing, bioinformatic characterization and expression pattern of a putative amino acid transporter from the parasitic cestode Echinococcus granulosus. Gene, 411(1–2), 1–9. - PubMed
-
- Collins PR, Stack CM, O’Neill SM, Doyle S, Ryan T, Brennan GP, Mousley A, Stewart M, Maule AG, Dalton JP, Donnelly S. 2004. Cathepsin L1, the major protease involved in liver fluke (Fasciola hepatica) virulence: propetide cleavage sites and autoactivation of the zymogen secreted from gastrodermal cells. Journal of Biological Chemistry, 279(17), 17038–17046. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
