IFIH1 (MDA5) is required for innate immune detection of intron-containing RNA expressed from the HIV-1 provirus
- PMID: 38985764
- PMCID: PMC11260138
- DOI: 10.1073/pnas.2404349121
IFIH1 (MDA5) is required for innate immune detection of intron-containing RNA expressed from the HIV-1 provirus
Abstract
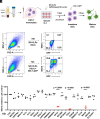
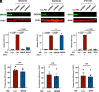
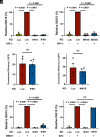
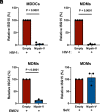
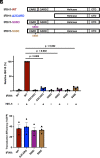
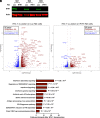
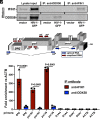
Intron-containing RNA expressed from the HIV-1 provirus activates type 1 interferon in primary human blood cells, including CD4+ T cells, macrophages, and dendritic cells. To identify the innate immune receptor required for detection of intron-containing RNA expressed from the HIV-1 provirus, a loss-of-function screen was performed with short hairpin RNA-expressing lentivectors targeting twenty-one candidate genes in human monocyte-derived dendritic cells. Among the candidate genes tested, only knockdown of XPO1 (CRM1), IFIH1 (MDA5), or MAVS prevented activation of the interferon-stimulated gene ISG15. The importance of IFIH1 protein was demonstrated by rescue of the knockdown with nontargetable IFIH1 coding sequence. Inhibition of HIV-1-induced ISG15 by the IFIH1-specific Nipah virus V protein, and by IFIH1-transdominant 2-CARD domain-deletion or phosphomimetic point mutations, indicates that IFIH1 (MDA5) filament formation, dephosphorylation, and association with MAVS are all required for innate immune activation in response to HIV-1 transduction. Since both IFIH1 (MDA5) and DDX58 (RIG-I) signal via MAVS, the specificity of HIV-1 RNA detection by IFIH1 was demonstrated by the fact that DDX58 knockdown had no effect on activation. RNA-Seq showed that IFIH1 knockdown in dendritic cells globally disrupted the induction of IFN-stimulated genes by HIV-1. Finally, specific enrichment of unspliced HIV-1 RNA by IFIH1 (MDA5), over two orders of magnitude, was revealed by formaldehyde cross-linking immunoprecipitation (f-CLIP). These results demonstrate that IFIH1 is the innate immune receptor for intron-containing RNA from the HIV-1 provirus and that IFIH1 potentially contributes to chronic inflammation in people living with HIV-1, even in the presence of effective antiretroviral therapy.
Keywords: HIV-1; MDA5; dendritic cells; type 1 interferon; unspliced RNA.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Update of
-
IFIH1 (MDA5) is required for innate immune detection of intron-containing RNA expressed from the HIV-1 provirus.bioRxiv [Preprint]. 2023 Dec 12:2023.11.17.567619. doi: 10.1101/2023.11.17.567619. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2404349121. doi: 10.1073/pnas.2404349121 PMID: 38014177 Free PMC article. Updated. Preprint.
Similar articles
-
IFIH1 (MDA5) is required for innate immune detection of intron-containing RNA expressed from the HIV-1 provirus.bioRxiv [Preprint]. 2023 Dec 12:2023.11.17.567619. doi: 10.1101/2023.11.17.567619. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2404349121. doi: 10.1073/pnas.2404349121 PMID: 38014177 Free PMC article. Updated. Preprint.
-
Complement Potentiates Immune Sensing of HIV-1 and Early Type I Interferon Responses.mBio. 2021 Oct 26;12(5):e0240821. doi: 10.1128/mBio.02408-21. Epub 2021 Oct 12. mBio. 2021. PMID: 34634939 Free PMC article.
-
Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS.J Virol. 2012 Aug;86(15):8245-58. doi: 10.1128/JVI.00215-12. Epub 2012 May 23. J Virol. 2012. PMID: 22623778 Free PMC article.
-
A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation.Trends Microbiol. 2019 Jan;27(1):75-85. doi: 10.1016/j.tim.2018.08.007. Epub 2018 Sep 7. Trends Microbiol. 2019. PMID: 30201512 Free PMC article. Review.
-
Negative regulators of the RIG-I-like receptor signaling pathway.Eur J Immunol. 2017 Apr;47(4):615-628. doi: 10.1002/eji.201646484. Eur J Immunol. 2017. PMID: 28295214 Free PMC article. Review.
Cited by
-
Help or Hinder: Protein Host Factors That Impact HIV-1 Replication.Viruses. 2024 Aug 10;16(8):1281. doi: 10.3390/v16081281. Viruses. 2024. PMID: 39205255 Free PMC article. Review.
References
-
- Wong J. K., et al. , Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). - PubMed
-
- Hunt P. W., et al. , T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J. Infect. Dis. 187, 1534–1543 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

