Systematic analysis of the expression profiles and prognostic significance of the MED gene family in renal clear cell carcinoma
- PMID: 38979551
- PMCID: PMC11228927
- DOI: 10.3892/ol.2024.14531
Systematic analysis of the expression profiles and prognostic significance of the MED gene family in renal clear cell carcinoma
Abstract
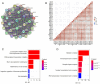
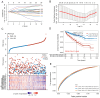
The mediator complex (MED) family is a contributing factor in the regulation of transcription and proliferation of cells, and is closely associated with the development of various types of cancer. However, the significance of the expression levels and prognostic value of MED genes in kidney renal clear cell carcinoma (KIRC) have rarely been reported. The present study analyzed the expression and prognostic potential of MED genes in KIRC. The Search Tool for the Retrieval of Interacting Genes/Proteins was used to construct the protein-protein interaction network (PPI), the Assistant for Clinical Bioinformatics database was used to perform correlation analysis, GEPIA 2 was utilized to draw the Kaplan-Meier plot and analyze prognostic significance and the Tumor Immune Estimation Resource was used to assess the association of MED genes with the infiltration of immune cells in patients with KIRC. A total of 30 MED genes were identified, and among these genes, 11 were selected for the creation of a prognostic gene signature based on the results of a LASSO Cox regression analysis. Furthermore, according to univariate and multivariate analyses, MED7, MED16, MED21, MED25 and MED29 may be valuable independent predictive biomarkers for the prognosis of individuals with KIRC. Furthermore, there were significant differences in the expression levels of MED7, MED21 and MED25 in KIRC among different tumor grades. Additionally, patients with KIRC with high transcription levels of MED7, MED21 and MED29 had considerably longer overall survival times. The expression levels of MED genes were also linked to the infiltration of several immune cells. Overall, MED genes may have potential significance in predicting the prognosis of patients with KIRC.
Keywords: MED gene family; bioinformatics analysis; renal clear cell carcinoma.
Copyright: © 2024 Wang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Systematic analysis of expression profiles and prognostic significance for MMDS-related iron-sulfur proteins in renal clear cell carcinoma.Sci Rep. 2022 Nov 16;12(1):19637. doi: 10.1038/s41598-022-22479-4. Sci Rep. 2022. PMID: 36385109 Free PMC article.
-
The clinical significance, immune infiltration, and tumor mutational burden of angiogenesis-associated lncRNAs in kidney renal clear cell carcinoma.Front Immunol. 2022 Jul 26;13:934387. doi: 10.3389/fimmu.2022.934387. eCollection 2022. Front Immunol. 2022. PMID: 35958561 Free PMC article.
-
Diagnostic and prognostic role of basic leucine zipper transcription factor in kidney renal clear cell carcinoma.Transl Androl Urol. 2022 Feb;11(2):238-252. doi: 10.21037/tau-21-1130. Transl Androl Urol. 2022. PMID: 35280662 Free PMC article.
-
A Novel Gene Signature of Tripartite Motif Family for Predicting the Prognosis in Kidney Renal Clear Cell Carcinoma and Its Association With Immune Cell Infiltration.Front Oncol. 2022 Mar 17;12:840410. doi: 10.3389/fonc.2022.840410. eCollection 2022. Front Oncol. 2022. PMID: 35371994 Free PMC article.
-
ZEB family is a prognostic biomarker and correlates with anoikis and immune infiltration in kidney renal clear cell carcinoma.BMC Med Genomics. 2024 Jun 5;17(1):153. doi: 10.1186/s12920-024-01895-7. BMC Med Genomics. 2024. PMID: 38840097 Free PMC article.
References
-
- Linehan WM, Schmidt LS, Crooks DR, Wei D, Srinivasan R, Lang M, Ricketts CJ. The metabolic basis of kidney cancer. Cancer Discov. 2019;9:1006–1021. doi: 10.1158/2159-8290.CD-18-1354. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
