HIV-protease inhibitors potentiate the activity of carfilzomib in triple-negative breast cancer
- PMID: 38969867
- PMCID: PMC11368961
- DOI: 10.1038/s41416-024-02774-9
HIV-protease inhibitors potentiate the activity of carfilzomib in triple-negative breast cancer
Abstract
Background: Resistance to chemotherapy is a major problem in the treatment of patients with triple-negative breast cancer (TNBC). Preclinical data suggest that TNBC is dependent on proteasomes; however, clinical observations indicate that the efficacy of proteasome inhibitors in TNBC may be limited, suggesting the need for combination therapies.
Methods: We compared bortezomib and carfilzomib and their combinations with nelfinavir and lopinavir in TNBC cell lines and primary cells with regard to their cytotoxic activity, functional proteasome inhibition, and induction of the unfolded protein response (UPR). Furthermore, we evaluated the involvement of sXBP1, ABCB1, and ABCG2 in the cytotoxic activity of drug combinations.
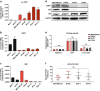
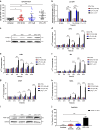
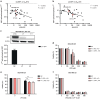
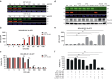
Results: Carfilzomib, via proteasome β5 + β2 inhibition, is more cytotoxic in TNBC than bortezomib, which inhibits β5 + β1 proteasome subunits. The cytotoxicity of carfilzomib was significantly potentiated by nelfinavir or lopinavir. Carfilzomib with lopinavir induced endoplasmic reticulum stress and pro-apoptotic UPR through the accumulation of excess proteasomal substrate protein in TNBC in vitro. Moreover, lopinavir increased the intracellular availability of carfilzomib by inhibiting carfilzomib export from cells that express high levels and activity of ABCB1, but not ABCG2.
Conclusion: Proteasome inhibition by carfilzomib combined with nelfinavir/lopinavir represents a potential treatment option for TNBC, warranting further investigation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Enhancement of Doxorubicin Efficacy by Bacopaside II in Triple-Negative Breast Cancer Cells.Biomolecules. 2025 Jan 3;15(1):55. doi: 10.3390/biom15010055. Biomolecules. 2025. PMID: 39858449 Free PMC article.
-
Multiomics analysis reveals the involvement of NET1 in tumour immune regulation and malignant progression.Sci Rep. 2025 Jan 2;15(1):56. doi: 10.1038/s41598-024-83714-8. Sci Rep. 2025. PMID: 39747410 Free PMC article.
-
Vodobatinib overcomes cancer multidrug resistance by attenuating the drug efflux function of ABCB1 and ABCG2.Eur J Pharmacol. 2025 Feb 5;988:177231. doi: 10.1016/j.ejphar.2024.177231. Epub 2024 Dec 24. Eur J Pharmacol. 2025. PMID: 39725134
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
A review on endoplasmic reticulum-dependent anti-breast cancer activity of herbal drugs: possible challenges and opportunities.J Drug Target. 2025 Feb;33(2):206-231. doi: 10.1080/1061186X.2024.2417189. Epub 2024 Oct 24. J Drug Target. 2025. PMID: 39404107 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

