Targeting IL-1 controls refractory pityriasis rubra pilaris
- PMID: 38959302
- PMCID: PMC11221491
- DOI: 10.1126/sciadv.ado2365
Targeting IL-1 controls refractory pityriasis rubra pilaris
Abstract
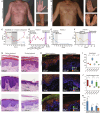
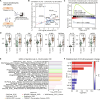
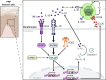
Pityriasis rubra pilaris (PRP) is a rare inflammatory skin disease with a poorly understood pathogenesis. Through a molecularly driven precision medicine approach and an extensive mechanistic pathway analysis in PRP skin samples, compared to psoriasis, atopic dermatitis, healed PRP, and healthy controls, we identified IL-1β as a key mediator, orchestrating an NF-κB-mediated IL-1β-CCL20 axis, including activation of CARD14 and NOD2. Treatment of three patients with the IL-1 antagonists anakinra and canakinumab resulted in rapid clinical improvement and reversal of the PRP-associated molecular signature with a 50% improvement in skin lesions after 2 to 3 weeks. This transcriptional signature was consistent with in vitro stimulation of keratinocytes with IL-1β. With the central role of IL-1β underscoring its potential as a therapeutic target, our findings propose a redefinition of PRP as an autoinflammatory keratinization disorder. Further clinical trials are needed to validate the efficacy of IL-1β antagonists in PRP.
Figures



Similar articles
-
Evaluation of Ixekizumab Treatment for Patients With Pityriasis Rubra Pilaris: A Single-Arm Trial.JAMA Dermatol. 2020 Jun 1;156(6):668-675. doi: 10.1001/jamadermatol.2020.0932. JAMA Dermatol. 2020. PMID: 32293641 Free PMC article. Clinical Trial.
-
Guselkumab for Pityriasis Rubra Pilaris and Dysregulation of IL-23/IL-17 and NFkB Signaling: A Nonrandomized Trial.JAMA Dermatol. 2024 Jun 1;160(6):641-645. doi: 10.1001/jamadermatol.2024.0257. JAMA Dermatol. 2024. PMID: 38598229 Clinical Trial.
-
CARD14 Glu138 mutation in familial pityriasis rubra pilaris does not warrant differentiation from familial psoriasis.J Dermatol. 2016 Feb;43(2):187-9. doi: 10.1111/1346-8138.13008. Epub 2015 Jul 1. J Dermatol. 2016. PMID: 26130407
-
CARD14-associated papulosquamous eruption (CAPE) in pediatric patients: Three additional cases and review of the literature.Pediatr Dermatol. 2021 Sep;38(5):1237-1242. doi: 10.1111/pde.14779. Epub 2021 Aug 26. Pediatr Dermatol. 2021. PMID: 34448248 Free PMC article. Review.
-
Autoinflammatory keratinization diseases: An emerging concept encompassing various inflammatory keratinization disorders of the skin.J Dermatol Sci. 2018 May;90(2):105-111. doi: 10.1016/j.jdermsci.2018.01.012. Epub 2018 Feb 1. J Dermatol Sci. 2018. PMID: 29422292 Review.
Cited by
-
Mutual Influence Between Allergic Rhinitis and Sleep: Factors, Mechanisms, and interventions-A Narrative Review.Nat Sci Sleep. 2024 Sep 19;16:1451-1467. doi: 10.2147/NSS.S482258. eCollection 2024. Nat Sci Sleep. 2024. PMID: 39318396 Free PMC article. Review.
References
-
- Kromer C., Sabat R., Celis D., Mössner R., Systemic therapies of pityriasis rubra pilaris: A systematic review. J. Dtsch. Dermatol. Ges. 17, 243–259 (2019). - PubMed
-
- Roenneberg S., Biedermann T., Pityriasis rubra pilaris: Algorithms for diagnosis and treatment. J. Eur. Acad. Dermatol. Venereol. 32, 889–898 (2018). - PubMed
-
- Boudreaux B. W., Pincelli T. P., Bhullar P. K., Patel M. H., Brumfiel C. M., Li X., Heckman M. G., Pittelkow M. R., Mangold A. R., Sluzevich J. C., Secukinumab for the treatment of adult-onset pityriasis rubra pilaris: A single-arm clinical trial with transcriptomic analysis. Br. J. Dermatol. 187, 650–658 (2022). - PubMed
-
- Iredale H. E., Meggitt S. J., Photosensitive pityriasis rubra pilaris. Clin. Exp. Dermatol. 31, 36–38 (2006). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

