Colletotrichum fructicola co-opts cytotoxic ribonucleases that antagonize host competitive microorganisms to promote infection
- PMID: 38953357
- PMCID: PMC11323725
- DOI: 10.1128/mbio.01053-24
Colletotrichum fructicola co-opts cytotoxic ribonucleases that antagonize host competitive microorganisms to promote infection
Abstract
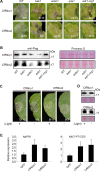
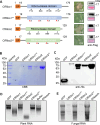
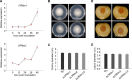
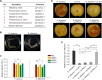
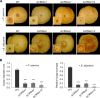
Phytopathogens secrete numerous molecules into the environment to establish a microbial niche and facilitate host infection. The phytopathogenic fungus Colletotrichum fructicola, which causes pear anthracnose, can colonize different plant tissues like leaves and fruits, which are occupied by a diversity of microbes. We speculate that this fungus produces antimicrobial effectors to outcompete host-associated competitive microorganisms. Herein, we identified two secreted ribonucleases, CfRibo1 and CfRibo2, from the C. fructicola secretome. The two ribonucleases both possess ribonuclease activity and showed cytotoxicity in Nicotianan benthamiana without triggering immunity in an enzymatic activity-dependent manner. CfRibo1 and CfRibo2 recombinant proteins exhibited toxicity against Escherichia coli, Saccharomyces cerevisiae, and, importantly, the phyllosphere microorganisms isolated from the pear host. Among these isolated microbial strains, Bacillus altitudinis is a pathogenic bacterium causing pear soft rot. Strikingly, CfRibo1 and CfRibo2 were found to directly antagonize B. altitudinis to facilitate C. fructicola infection. More importantly, CfRibo1 and CfRibo2 functioned as essential virulence factors of C. fructicola in the presence of host-associated microorganisms. Further analysis revealed these two ribonucleases are widely distributed in fungi and are undergoing purifying selection. Our results provide the first evidence of antimicrobial effectors in Colletotrichum fungi and extend the functional diversity of fungal ribonucleases in plant-pest-environment interactions.
Importance: Colletotrichum fructicola is emerging as a devastating pathogenic fungus causing anthracnose in various crops in agriculture, and understanding how this fungus establishes successful infection is of great significance for anthracnose disease management. Fungi are known to produce secreted effectors as weapons to promote virulence. Considerable progress has been made in elucidating how effectors manipulate plant immunity; however, their importance in modulating environmental microbes is frequently neglected. The present study identified two secreted ribonucleases, CfRibo1 and CfRibo2, as antimicrobial effectors of C. fructicola. These two proteins both possess toxicity to pear phyllosphere microorganisms, and they efficiently antagonize competitive microbes to facilitate the infection of pear hosts. This study represents the first evidence of antimicrobial effectors in Colletotrichum fungi, and we consider that CfRibo1 and CfRibo2 could be targeted for anthracnose disease management in diverse crops in the future.
Keywords: Colletotrichum; antimicrobial activity; cell death; host-associated microorganisms; ribonuclease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

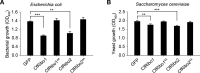

Similar articles
-
Extracellular perception of multiple novel core effectors from the broad host-range pear anthracnose pathogen Colletotrichum fructicola in the nonhost Nicotiana benthamiana.Hortic Res. 2024 Mar 14;11(5):uhae078. doi: 10.1093/hr/uhae078. eCollection 2024 May. Hortic Res. 2024. PMID: 38766536 Free PMC article.
-
A novel effector CfEC92 of Colletotrichum fructicola contributes to glomerella leaf spot virulence by suppressing plant defences at the early infection phase.Mol Plant Pathol. 2020 Jul;21(7):936-950. doi: 10.1111/mpp.12940. Epub 2020 Apr 22. Mol Plant Pathol. 2020. PMID: 32512647 Free PMC article.
-
Transcriptomic analysis reveals candidate genes regulating development and host interactions of Colletotrichum fructicola.BMC Genomics. 2018 Jul 28;19(1):557. doi: 10.1186/s12864-018-4934-0. BMC Genomics. 2018. PMID: 30055574 Free PMC article.
-
The Role of Virulence Factors in the Pathogenicity of Colletotrichum sp.Curr Protein Pept Sci. 2017;18(10):1005-1018. doi: 10.2174/1389203717666160813160727. Curr Protein Pept Sci. 2017. PMID: 27526925 Review.
-
Soybean anthracnose caused by Colletotrichum species: Current status and future prospects.Mol Plant Pathol. 2021 Apr;22(4):393-409. doi: 10.1111/mpp.13036. Epub 2021 Feb 20. Mol Plant Pathol. 2021. PMID: 33609073 Free PMC article. Review.
References
-
- Snelders NC, Boshoven JC, Song Y, Schmitz N, Fiorin GL, Rovenich H, van den Berg GCM, Torres DE, Petti GC, Prockl Z, Faino L, Seidl MF, Thomma B. 2023. A highly polymorphic effector protein promotes fungal virulence through suppression of plant‐associated Actinobacteria. New Phytol 237:944–958. doi:10.1111/nph.18576 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
