Evidence for widespread translation of 5' untranslated regions
- PMID: 38953162
- PMCID: PMC11317171
- DOI: 10.1093/nar/gkae571
Evidence for widespread translation of 5' untranslated regions
Abstract
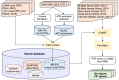
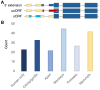
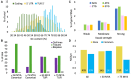
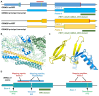
Ribosome profiling experiments support the translation of a range of novel human open reading frames. By contrast, most peptides from large-scale proteomics experiments derive from just one source, 5' untranslated regions. Across the human genome we find evidence for 192 translated upstream regions, most of which would produce protein isoforms with extended N-terminal ends. Almost all of these N-terminal extensions are from highly abundant genes, which suggests that the novel regions we detect are just the tip of the iceberg. These upstream regions have characteristics that are not typical of coding exons. Their GC-content is remarkably high, even higher than 5' regions in other genes, and a large majority have non-canonical start codons. Although some novel upstream regions have cross-species conservation - five have orthologues in invertebrates for example - the reading frames of two thirds are not conserved beyond simians. These non-conserved regions also have no evidence of purifying selection, which suggests that much of this translation is not functional. In addition, non-conserved upstream regions have significantly more peptides in cancer cell lines than would be expected, a strong indication that an aberrant or noisy translation initiation process may play an important role in translation from upstream regions.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Diversity of translation start sites may define increased complexity of the human short ORFeome.Mol Cell Proteomics. 2007 Jun;6(6):1000-6. doi: 10.1074/mcp.M600297-MCP200. Epub 2007 Feb 21. Mol Cell Proteomics. 2007. PMID: 17317662
-
Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides.BMC Genomics. 2006 Jan 26;7:16. doi: 10.1186/1471-2164-7-16. BMC Genomics. 2006. PMID: 16438715 Free PMC article.
-
Genome-wide identification of Arabidopsis non-AUG-initiated upstream ORFs with evolutionarily conserved regulatory sequences that control protein expression levels.Plant Mol Biol. 2023 Jan;111(1-2):37-55. doi: 10.1007/s11103-022-01309-1. Epub 2022 Aug 31. Plant Mol Biol. 2023. PMID: 36044152
-
The interplay between cis- and trans-acting factors drives selective mRNA translation initiation in eukaryotes.Biochimie. 2024 Feb;217:20-30. doi: 10.1016/j.biochi.2023.09.017. Epub 2023 Sep 21. Biochimie. 2024. PMID: 37741547 Review.
-
Translational control by 5'-untranslated regions of eukaryotic mRNAs.Science. 2016 Jun 17;352(6292):1413-6. doi: 10.1126/science.aad9868. Science. 2016. PMID: 27313038 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

