Targeting dermatophyte Cdc42 and Rac GTPase signaling to hinder hyphal elongation and virulence
- PMID: 38952678
- PMCID: PMC11215307
- DOI: 10.1016/j.isci.2024.110139
Targeting dermatophyte Cdc42 and Rac GTPase signaling to hinder hyphal elongation and virulence
Abstract
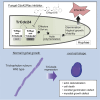
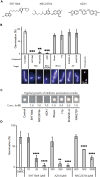
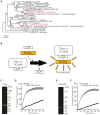
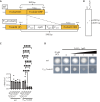
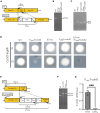
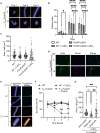
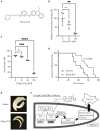
The development of antifungal drugs requires novel molecular targets due to limited treatment options and drug resistance. Through chemical screening and establishment of a novel genetic technique to repress gene expression in Trichophyton rubrum, the primary causal fungus of dermatophytosis, we demonstrated that fungal Cdc42 and Rac GTPases are promising antifungal drug targets. Chemical inhibitors of these GTPases impair hyphal formation, which is crucial for growth and virulence in T. rubrum. Conditional repression of Cdc24, a guanine nucleotide exchange factor for Cdc42 and Rac, led to hyphal growth defects, abnormal cell morphology, and cell death. EHop-016 inhibited the promotion of the guanine nucleotide exchange reaction in Cdc42 and Rac by Cdc24 as well as germination and growth on the nail fragments of T. rubrum and improved animal survival in an invertebrate infection model of T. rubrum. Our results provide a novel antifungal therapeutic target and a potential lead compound.
Keywords: Microbiology; Mycology; cell biology; molecular biology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth.Eukaryot Cell. 2005 Mar;4(3):588-603. doi: 10.1128/EC.4.3.588-603.2005. Eukaryot Cell. 2005. PMID: 15755921 Free PMC article.
-
Rac and Cdc42 inhibitors reduce macrophage function in breast cancer preclinical models.Front Oncol. 2023 Jun 16;13:1152458. doi: 10.3389/fonc.2023.1152458. eCollection 2023. Front Oncol. 2023. PMID: 37397366 Free PMC article.
-
Roles of Aspergillus nidulans Cdc42/Rho GTPase regulators in hyphal morphogenesis and development.Mycologia. 2016 May-Jun;108(3):543-55. doi: 10.3852/15-232. Epub 2016 Mar 1. Mycologia. 2016. PMID: 26932184
-
Targeting Rac and Cdc42 GEFs in Metastatic Cancer.Front Cell Dev Biol. 2020 Apr 8;8:201. doi: 10.3389/fcell.2020.00201. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32322580 Free PMC article. Review.
-
Targeting Rac and Cdc42 GTPases in Cancer.Cancer Res. 2018 Jun 15;78(12):3101-3111. doi: 10.1158/0008-5472.CAN-18-0619. Epub 2018 Jun 1. Cancer Res. 2018. PMID: 29858187 Free PMC article. Review.
Cited by
-
An efficient gene targeting system using Δku80 and functional analysis of Cyp51A in Trichophyton rubrum.AMB Express. 2024 Aug 31;14(1):96. doi: 10.1186/s13568-024-01755-8. AMB Express. 2024. PMID: 39215862 Free PMC article.
References
-
- Hayette M.-P., Sacheli R. Dermatophytosis, Trends in Epidemiology and Diagnostic Approach. Curr. Fungal Infect. Rep. 2015;9:164–179. doi: 10.1007/s12281-015-0231-4. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

