This is a preprint.
Precise measurement of molecular phenotypes with barcode-based CRISPRi systems
- PMID: 38948701
- PMCID: PMC11213135
- DOI: 10.1101/2024.06.21.600132
Precise measurement of molecular phenotypes with barcode-based CRISPRi systems
Abstract
Genome-wide CRISPR-Cas9 screens have untangled regulatory networks and revealed the genetic underpinnings of diverse biological processes. Their success relies on experimental designs that interrogate specific molecular phenotypes and distinguish key regulators from background effects. Here, we realize these goals with a generalizable platform for CRISPR interference with barcoded expression reporter sequencing (CiBER-seq) that dramatically improves the sensitivity and scope of genome-wide screens. We systematically address technical factors that distort phenotypic measurements by normalizing expression reporters against closely-matched control promoters, integrated together into the genome at single copy. To test our ability to capture post-transcriptional and post-translational regulation through sequencing, we screened for genes that affected nonsense-mediated mRNA decay and Doa10-mediated cytosolic protein decay. Our optimized CiBER-seq screens accurately capture the known components of well-studied RNA and protein quality control pathways with minimal background. These results demonstrate the precision and versatility of CiBER-seq for dissecting the genetic networks controlling cellular behaviors.
Conflict of interest statement
Declaration of interests: N.T.I holds equity and serves as a scientific advisor to Tevard Biosciences, and holds equity in Velia Therapeutics.
Figures
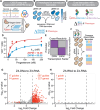
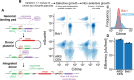
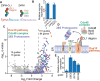

Similar articles
-
CRISPRi with barcoded expression reporters dissects regulatory networks in human cells.bioRxiv [Preprint]. 2024 Sep 6:2024.09.06.611573. doi: 10.1101/2024.09.06.611573. bioRxiv. 2024. PMID: 39282439 Free PMC article. Preprint.
-
Plasmid and Sequencing Library Preparation for CRISPRi Barcoded Expression Reporter Sequencing (CiBER-seq) in Saccharomyces cerevisiae.Bio Protoc. 2022 Apr 5;12(7):e4376. doi: 10.21769/BioProtoc.4376. eCollection 2022 Apr 5. Bio Protoc. 2022. PMID: 35530514 Free PMC article.
-
CiBER-seq dissects genetic networks by quantitative CRISPRi profiling of expression phenotypes.Science. 2020 Dec 11;370(6522):eabb9662. doi: 10.1126/science.abb9662. Science. 2020. PMID: 33303588 Free PMC article.
-
New Developments in CRISPR/Cas-based Functional Genomics and their Implications for Research Using Zebrafish.Curr Gene Ther. 2017;17(4):286-300. doi: 10.2174/1566523217666171121164132. Curr Gene Ther. 2017. PMID: 29173171 Review.
-
CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine.ACS Chem Biol. 2018 Feb 16;13(2):406-416. doi: 10.1021/acschembio.7b00657. Epub 2017 Oct 24. ACS Chem Biol. 2018. PMID: 29035510 Free PMC article. Review.
References
-
- Przybyla L. & Gilbert L. A. A new era in functional genomics screens. Nat. Rev. Genet. 23, 89–103 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
