This is a preprint.
Alzheimer's disease CSF biomarkers correlate with early pathology and alterations in neuronal and glial gene expression
- PMID: 38947015
- PMCID: PMC11213077
- DOI: 10.1101/2024.06.11.24308706
Alzheimer's disease CSF biomarkers correlate with early pathology and alterations in neuronal and glial gene expression
Update in
-
Alzheimer's disease CSF biomarkers correlate with early pathology and alterations in neuronal and glial gene expression.Alzheimers Dement. 2024 Oct;20(10):7090-7103. doi: 10.1002/alz.14194. Epub 2024 Aug 27. Alzheimers Dement. 2024. PMID: 39192661 Free PMC article.
Abstract
Introduction: Normal pressure hydrocephalus (NPH) patients undergoing cortical shunting frequently show early AD pathology on cortical biopsy, which is predictive of progression to clinical AD. The objective of this study was to use samples from this cohort to identify CSF biomarkers for AD-related CNS pathophysiologic changes using tissue and fluids with early pathology, free of post-mortem artifact.
Methods: We analyzed Simoa, proteomic, and metabolomic CSF data from 81 patients with previously documented pathologic and transcriptomic changes.
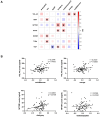
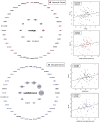
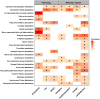
Results: AD pathology on biopsy correlates with CSF β-amyloid-40/42, neurofilament light chain (NfL), and phospho-tau-181(p-tau181)/β-amyloid-42, while several gene expression modules correlate with NfL. Proteomic analysis highlights 7 core proteins that correlate with pathology and gene expression changes on biopsy, and metabolomic analysis of CSF identifies disease-relevant groups that correlate with biopsy data..
Discussion: As additional biomarkers are added to AD diagnostic panels, our work provides insight into the CNS pathophysiology these markers are tracking.
Keywords: Alzheimer’s disease; CSF; biomarkers; metabolomics; proteomics.
Conflict of interest statement
Declaration of Interests/Conflicts: LSH reports grants from NIH, New York State Dept of Health, Lewy Body Disease Association, CurePSP, Abbvie, Acumen, Alector, Biogen, Bristol-Myer Squibb, Cognition, EIP, Eisai, Genentech/Roche, Janssen/Johnson and Johnson, Transposon Therapeutics, UCB, and Vaccinex, as well as consulting fees from Biogen, Corium, Eisai, Genentech/Roche, and New Amsterdam, Payment or honoraria from Eisai Pharmaceuticals, Medscape, and Biogen, Payment for expert testimony from Monsanto and legal firms, support for attending meetings and/or travel from Eisai Pharmaceuticals, participation on a data safety monitoring or advisory board from Prevail Therapeutics/Lilly, Cortexyme, and Eisai, and a leadership role in the Alzheimer’s Association. GWM reports grant funding from NIH, CancerUK, Department of Defense (USAMRAA), Alley Corp, and SPARK-NS. AFT reports grant funding from NIH and Regeneron, stock ownership in Biogen and Ionis, paid committee work for DOD and NIH, and unpaid committee work for the Alzheimer’s Association. RAM reports grants from NIH, Minnesota Partnership for Biotechnology and Genomics, Minnesota Robotics Institute, and MnDRIVE Data Science Initiative. GMM reports grants from NIH, consulting with Koh Young Inc and NeuroOne Technologies, participation on the Medronic SLATE trial Publication Committee, and leadership/committee roles in the Neurosurgical Society of America, AANS, and ASSFN. LMB reports support from NIH. The other authors have nothing to report.
Figures

Similar articles
-
Alzheimer's disease CSF biomarkers correlate with early pathology and alterations in neuronal and glial gene expression.Alzheimers Dement. 2024 Oct;20(10):7090-7103. doi: 10.1002/alz.14194. Epub 2024 Aug 27. Alzheimers Dement. 2024. PMID: 39192661 Free PMC article.
-
Plasma neurofilament light and phosphorylated tau 181 as biomarkers of Alzheimer's disease pathology and clinical disease progression.Alzheimers Res Ther. 2021 Mar 25;13(1):65. doi: 10.1186/s13195-021-00805-8. Alzheimers Res Ther. 2021. PMID: 33766131 Free PMC article.
-
Time Trends of Cerebrospinal Fluid Biomarkers of Neurodegeneration in Idiopathic Normal Pressure Hydrocephalus.J Alzheimers Dis. 2021;80(4):1629-1642. doi: 10.3233/JAD-201361. J Alzheimers Dis. 2021. PMID: 33720890 Free PMC article.
-
Core candidate neurochemical and imaging biomarkers of Alzheimer's disease.Alzheimers Dement. 2008 Jan;4(1):38-48. doi: 10.1016/j.jalz.2007.08.006. Epub 2007 Dec 21. Alzheimers Dement. 2008. PMID: 18631949 Review.
-
A Review of Fluid Biomarkers for Alzheimer's Disease: Moving from CSF to Blood.Neurol Ther. 2017 Jul;6(Suppl 1):15-24. doi: 10.1007/s40120-017-0073-9. Epub 2017 Jul 21. Neurol Ther. 2017. PMID: 28733960 Free PMC article. Review.
References
-
- Borgesen SE, Gjerris F. The predictive value of conductance to outflow of CSF in normal pressure hydrocephalus. Brain. 1982;105:65–86. - PubMed
-
- Symon L, Dorsch NW. Use of long-term intracranial pressure measurement to assess hydrocephalic patients prior to shunt surgery. Journal of neurosurgery. 1975;42:258–73. - PubMed
-
- McGovern RA, Nelp TB, Kelly KM, Chan AK, Mazzoni P, Sheth SA, et al. Predicting Cognitive Improvement in Normal Pressure Hydrocephalus Patients Using Preoperative Neuropsychological Testing and Cerebrospinal Fluid Biomarkers. Neurosurgery. 2019;85:E662–E9. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
