FUT11 expression in gastric cancer: its prognostic significance and role in immune regulation
- PMID: 38941002
- PMCID: PMC11213843
- DOI: 10.1007/s12672-024-01120-y
FUT11 expression in gastric cancer: its prognostic significance and role in immune regulation
Abstract
Background: Gastric cancer (GC) is a malignant digestive tract tumor with a high recurrence rate and poor prognosis. Fucosylation is important in tumor glycosylation, in which the key enzyme is fucosyltransferase (FUT). FUT11 is a member of the fucosyltransferase family and has been closely associated with the development of multiple cancers. However, the specific relationship between FUT11 and GC prognosis and its molecular mechanism has not been fully studied. This study explored FUT11 expression, clinical correlation, and its role in GC occurrence and development to deepen understanding of its function.
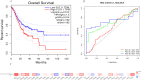
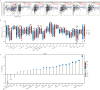
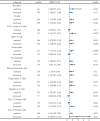
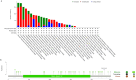
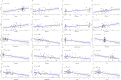
Methods: FUT11 expression in 33 cancers was preliminarily analyzed using the Tumor Immunoassay Resource (TIMER2.0) database. FUT11 expression in GC was evaluated using The Cancer Genome Atlas stomach adenocarcinoma (TCGA-STAD) and Gene Expression Profiling Interactive Analysis (GEPIA2) data and verified using the Gene Expression Omnibus (GEO) GSE65801 dataset. Furthermore, we studied the survival prognosis of FUT11 in GC and analyzed its effect on the survival rate of patients with GC using the KM-plotter. We also performed COX regression analysis on TCGA GC clinical data and analyzed FUT11 expression in the pathway using the STRING and LinkedOmics databases. Moreover, the relationship between FUT11 and GC immune infiltration level was examined, and the Kaplan-Meier survival analysis diagram was constructed. The FUT11 genetic variation information was retrieved using cBioPortal, and its drug sensitivity was analyzed using CellMiner. Finally, differential FUT11 expression in GC tissues was verified using immunohistochemistry.
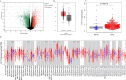
Results: The data mining and analysis demonstrated that FUT11 expression was abnormally elevated in GC tissues and correlated with poor patient prognosis. The FUT11 expression level was an independent prognostic factor for GC. The difference in FUT11 expression level resulted in different degrees of immune cell infiltration in the patients with GC, which might regulate the tumor microenvironment. FUT11 affected GC development by participating in cancer pathways such as PI3K-AKT, neuroactive ligand-receptor, and MAPK. Immunohistochemical staining revealed that FUT11 was highly expressed in GC.
Conclusions: This study revealed that FUT11 expression is significantly increased in GC tissues. This increase is associated with poor prognosis and might affect immune regulation. FUT11 might have immunological and targeted therapeutic value, providing a new approach to GC treatment.
Keywords: Bioinformatics; FUT11; Fucosylation; GC; Immunology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Knockdown of FUT11 inhibits the progression of gastric cancer via the PI3K/AKT pathway.Heliyon. 2023 Jul 7;9(7):e17600. doi: 10.1016/j.heliyon.2023.e17600. eCollection 2023 Jul. Heliyon. 2023. PMID: 37483811 Free PMC article.
-
Comprehensive bioinformatic analysis of mind bomb 1 gene in stomach adenocarcinoma.World J Gastrointest Oncol. 2023 Jul 15;15(7):1295-1310. doi: 10.4251/wjgo.v15.i7.1295. World J Gastrointest Oncol. 2023. PMID: 37546549 Free PMC article.
-
High expression of TMEM200A is associated with a poor prognosis and immune infiltration in gastric cancer.Pathol Oncol Res. 2023 Jan 19;29:1610893. doi: 10.3389/pore.2023.1610893. eCollection 2023. Pathol Oncol Res. 2023. PMID: 36741965 Free PMC article.
-
A novel necroptosis-related gene index for predicting prognosis and a cold tumor immune microenvironment in stomach adenocarcinoma.Front Immunol. 2022 Oct 27;13:968165. doi: 10.3389/fimmu.2022.968165. eCollection 2022. Front Immunol. 2022. PMID: 36389725 Free PMC article. Review.
-
Systems biology and OMIC data integration to understand gastrointestinal cancers.World J Clin Oncol. 2022 Oct 24;13(10):762-778. doi: 10.5306/wjco.v13.i10.762. World J Clin Oncol. 2022. PMID: 36337313 Free PMC article. Review.
Cited by
-
Genomic analyses identify 15 susceptibility loci and reveal HDAC2, SOX2-OT, and IGF2BP2 in a naturally-occurring canine model of gastric cancer.bioRxiv [Preprint]. 2024 Aug 16:2024.08.14.604426. doi: 10.1101/2024.08.14.604426. bioRxiv. 2024. PMID: 39372775 Free PMC article. Preprint.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
