High Polymorphism Levels of De Novo ORFs in a Yoruba Human Population
- PMID: 38934859
- PMCID: PMC11221430
- DOI: 10.1093/gbe/evae126
High Polymorphism Levels of De Novo ORFs in a Yoruba Human Population
Abstract
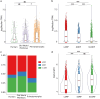
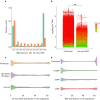
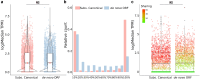
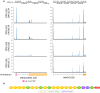
During evolution, new open reading frames (ORFs) with the potential to give rise to novel proteins continuously emerge. A recent compilation of noncanonical ORFs with translation signatures in humans has identified thousands of cases with a putative de novo origin. However, it is not known which is their distribution in the population. Are they universally translated? Here, we use ribosome profiling data from 65 lymphoblastoid cell lines from individuals of Yoruba origin to investigate this question. We identify 2,587 de novo ORFs translated in at least one of the cell lines. In line with their de novo origin, the encoded proteins tend to be smaller than 100 amino acids and encode positively charged proteins. We observe that the de novo ORFs are more polymorphic in the population than the set of canonical proteins, with a substantial fraction of them being translated in only some of the cell lines. Remarkably, this difference remains significant after controlling for differences in the translation levels. These results suggest that variations in the level translation of de novo ORFs could be a relevant source of intraspecies phenotypic diversity in humans.
Keywords: LCL; de novo ORF; human; polymorphism; translation.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Translation of Small Open Reading Frames: Roles in Regulation and Evolutionary Innovation.Trends Genet. 2019 Mar;35(3):186-198. doi: 10.1016/j.tig.2018.12.003. Epub 2018 Dec 31. Trends Genet. 2019. PMID: 30606460 Review.
-
Turnover of ribosome-associated transcripts from de novo ORFs produces gene-like characteristics available for de novo gene emergence in wild yeast populations.Genome Res. 2019 Jun;29(6):932-943. doi: 10.1101/gr.239822.118. Epub 2019 May 31. Genome Res. 2019. PMID: 31152050 Free PMC article.
-
Evolution of new proteins from translated sORFs in long non-coding RNAs.Exp Cell Res. 2020 Jun 1;391(1):111940. doi: 10.1016/j.yexcr.2020.111940. Epub 2020 Mar 7. Exp Cell Res. 2020. PMID: 32156600 Review.
-
De novo Identification of Actively Translated Open Reading Frames with Ribosome Profiling Data.J Vis Exp. 2022 Feb 18;(180). doi: 10.3791/63366. J Vis Exp. 2022. PMID: 35253791
-
Detecting actively translated open reading frames in ribosome profiling data.Nat Methods. 2016 Feb;13(2):165-70. doi: 10.1038/nmeth.3688. Epub 2015 Dec 14. Nat Methods. 2016. PMID: 26657557
References
-
- Charif D, Lobry JR. Seqinr 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Structural approaches to sequence evolution: molecules, networks, populations. New York: Biological and Medical Physics, Biomedical Engineering Springer Verlag; 2007. p. 207–232.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

