A Molecular Characterization of the Allelic Expression of the BRCA1 Founder Δ9-12 Pathogenic Variant and Its Potential Clinical Relevance in Hereditary Cancer
- PMID: 38928478
- PMCID: PMC11204022
- DOI: 10.3390/ijms25126773
A Molecular Characterization of the Allelic Expression of the BRCA1 Founder Δ9-12 Pathogenic Variant and Its Potential Clinical Relevance in Hereditary Cancer
Abstract
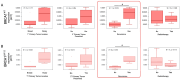
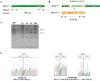
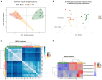
Hereditary breast and ovarian cancer (HBOC) syndrome is a genetic condition that increases the risk of breast cancer by 80% and that of ovarian cancer by 40%. The most common pathogenic variants (PVs) causing HBOC occur in the BRCA1 gene, with more than 3850 reported mutations in the gene sequence. The prevalence of specific PVs in BRCA1 has increased across populations due to the effect of founder mutations. Therefore, when a founder mutation is identified, it becomes key to improving cancer risk characterization and effective screening protocols. The only founder mutation described in the Mexican population is the deletion of exons 9 to 12 of BRCA1 (BRCA1Δ9-12), and its description focuses on the gene sequence, but no transcription profiles have been generated for individuals who carry this gene. In this study, we describe the transcription profiles of cancer patients and healthy individuals who were heterozygous for PV BRCA1Δ9-12 by analyzing the differential expression of both alleles compared with the homozygous BRCA1 control group using RT-qPCR, and we describe the isoforms produced by the BRCA1 wild-type and BRCA1Δ9-12 alleles using nanopore long-sequencing. Using the Kruskal-Wallis test, our results showed a similar transcript expression of the wild-type allele between the healthy heterozygous group and the homozygous BRCA1 control group. An association between the recurrence and increased expression of both alleles in HBOC patients was also observed. An analysis of the sequences indicated four wild-type isoforms with diagnostic potential for discerning individuals who carry the PV BRCA1Δ9-12 and identifying which of them has developed cancer.
Keywords: BRCA1; BRCA1Δ9–12; allele differential expression; founder mutation; hereditary breast and ovarian cancer; isoform; nanopore sequencing; pathogenic variants.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Germline mutational variants of Turkish ovarian cancer patients suspected of Hereditary Breast and Ovarian Cancer (HBOC) by next-generation sequencing.Pathol Res Pract. 2024 Feb;254:155075. doi: 10.1016/j.prp.2023.155075. Epub 2024 Jan 2. Pathol Res Pract. 2024. PMID: 38219492
-
Mexican BRCA1 founder mutation: Shortening the gap in genetic assessment for hereditary breast and ovarian cancer patients.PLoS One. 2019 Sep 23;14(9):e0222709. doi: 10.1371/journal.pone.0222709. eCollection 2019. PLoS One. 2019. PMID: 31545835 Free PMC article.
-
Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals.Breast Cancer Res. 2011 Feb 28;13(1):R20. doi: 10.1186/bcr2832. Breast Cancer Res. 2011. PMID: 21356067 Free PMC article.
-
The role of BRCA1/2 in hereditary and familial breast and ovarian cancers.Mol Genet Genomic Med. 2019 Sep;7(9):e879. doi: 10.1002/mgg3.879. Epub 2019 Jul 17. Mol Genet Genomic Med. 2019. PMID: 31317679 Free PMC article. Review.
-
Peritoneal carcinoma in women with genetic susceptibility: implications for Jewish populations.Fam Cancer. 2004;3(3-4):265-81. doi: 10.1007/s10689-004-9554-y. Fam Cancer. 2004. PMID: 15516851 Review.
References
-
- Villarreal-Garza C., Alvarez-Gómez R.M., Pérez-Plasencia C., Herrera L.A., Herzog J., Castillo D., Mohar A., Castro C., Gallardo L.N., Gallardo D., et al. Significant Clinical Impact of Recurrent BRCA1 and BRCA2 Mutations in Mexico. Cancer. 2015;121:372–378. doi: 10.1002/cncr.29058. - DOI - PMC - PubMed
-
- Daly M.B., Pal T., Berry M.P., Buys S.S., Dickson P., Domchek S.M., Elkhanany A., Friedman S., Goggins M., Hutton M.L., et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021;19:77–102. doi: 10.6004/jnccn.2021.0001. - DOI - PubMed
-
- Torres-Mejía G., Royer R., Llacuachaqui M., Akbari M.R., Giuliano A.R., Martínez-Matsushita L., Angeles-Llerenas A., Ortega-Olvera C., Ziv E., Lazcano-Ponce E., et al. Recurrent BRCA1 and BRCA2 Mutations in Mexican Women with Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2015;24:498–505. doi: 10.1158/1055-9965.EPI-13-0980. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

