Chloroplast Genomes of Vitis flexuosa and Vitis amurensis: Molecular Structure, Phylogenetic, and Comparative Analyses for Wild Plant Conservation
- PMID: 38927697
- PMCID: PMC11203327
- DOI: 10.3390/genes15060761
Chloroplast Genomes of Vitis flexuosa and Vitis amurensis: Molecular Structure, Phylogenetic, and Comparative Analyses for Wild Plant Conservation
Abstract
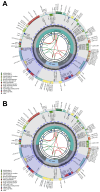
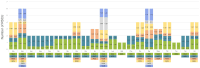
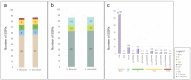
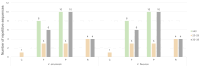
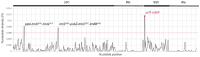
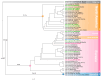
The chloroplast genome plays a crucial role in elucidating genetic diversity and phylogenetic relationships. Vitis vinifera L. (grapevine) is an economically important species, prompting exploration of wild genetic resources to enhance stress resilience. We meticulously assembled the chloroplast genomes of two Korean Vitis L. species, V. flexuosa Thunb. and V. amurensis Rupr., contributing valuable data to the Korea Crop Wild Relatives inventory. Through exhaustive specimen collection spanning diverse ecological niches across South Korea, we ensured comprehensive representation of genetic diversity. Our analysis, which included rigorous codon usage bias assessment and repeat analysis, provides valuable insights into amino acid preferences and facilitates the identification of potential molecular markers. The assembled chloroplast genomes were subjected to meticulous annotation, revealing divergence hotspots enriched with nucleotide diversity, thereby presenting promising candidates for DNA barcodes. Additionally, phylogenetic analysis reaffirmed intra-genus relationships and identified related crops, shedding light on evolutionary patterns within the genus. Comparative examination with chloroplast genomes of other crops uncovered conserved sequences and variable regions, offering critical insights into genetic evolution and adaptation. Our study advances the understanding of chloroplast genomes, genetic diversity, and phylogenetic relationships within Vitis species, thereby laying a foundation for enhancing grapevine genetic diversity and resilience to environmental challenges.
Keywords: Vitis; chloroplast; conservation; genetic diversity; genome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Chloroplast Genome Variation and Phylogenetic Relationships of Autochthonous Varieties of Vitis vinifera from the Don Valley.Int J Mol Sci. 2024 Sep 14;25(18):9928. doi: 10.3390/ijms25189928. Int J Mol Sci. 2024. PMID: 39337416 Free PMC article.
-
Comparative Chloroplast Genomes Analysis Provided Adaptive Evolution Insights in Medicago ruthenica.Int J Mol Sci. 2024 Aug 9;25(16):8689. doi: 10.3390/ijms25168689. Int J Mol Sci. 2024. PMID: 39201375 Free PMC article.
-
Decoding the Chloroplast Genome of Tetrastigma (Vitaceae): Variations and Phylogenetic Selection Insights.Int J Mol Sci. 2024 Jul 29;25(15):8290. doi: 10.3390/ijms25158290. Int J Mol Sci. 2024. PMID: 39125860 Free PMC article.
-
Chloroplast genomes: diversity, evolution, and applications in genetic engineering.Genome Biol. 2016 Jun 23;17(1):134. doi: 10.1186/s13059-016-1004-2. Genome Biol. 2016. PMID: 27339192 Free PMC article. Review.
-
The wild side of grape genomics.Trends Genet. 2024 Jul;40(7):601-612. doi: 10.1016/j.tig.2024.04.014. Epub 2024 May 21. Trends Genet. 2024. PMID: 38777691 Review.
References
-
- Jarvis A., Lane A., Hijmans R.J. The effect of climate change on crop wild relatives. Agric. Ecosyst. Environ. 2008;126:13–23. doi: 10.1016/j.agee.2008.01.013. - DOI
-
- Coyne C.J., Kumar S., von Wettberg E.J., Marques E., Berger J.D., Redden R.J., Ellis T.N., Brus J., Zablatzká L., Smýkal P. Potential and limits of exploitation of crop wild relatives for pea, lentil, and chickpea improvement. Legume Sci. 2020;2:e36. doi: 10.1002/leg3.36. - DOI
-
- Wen J., Lu L.M., Nie Z.L., Liu X.Q., Zhang N., Ickert-Bond S., Gerrath J., Manchester S.R., Boggan J., Chen Z.D. A new phylogenetic tribal classification of the grape family (Vitaceae) J. Syst. Evol. 2018;56:262–272. doi: 10.1111/jse.12427. - DOI
-
- Wan Y., Schwaninger H.R., Baldo A.M., Labate J.A., Zhong G.-Y., Simon C.J. A phylogenetic analysis of the grape genus (Vitis L.) reveals broad reticulation and concurrent diversification during neogene and quaternary climate change. BMC Evol. Biol. 2013;13:141. doi: 10.1186/1471-2148-13-141. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

