Lack of mismatch repair enhances resistance to methylating agents for cells deficient in oxidative demethylation
- PMID: 38925328
- PMCID: PMC11326903
- DOI: 10.1016/j.jbc.2024.107492
Lack of mismatch repair enhances resistance to methylating agents for cells deficient in oxidative demethylation
Abstract
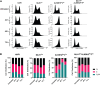
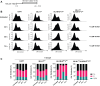
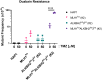
The human alkylation B (AlkB) homologs, ALKBH2 and ALKBH3, respond to methylation damage to maintain genomic integrity and cellular viability. Both ALKBH2 and ALKBH3 are direct reversal repair enzymes that remove 1-methyladenine (1meA) and 3-methylcytosine (3meC) lesions commonly generated by alkylating chemotherapeutic agents. Thus, the existence of deficiencies in ALKBH proteins can be exploited in synergy with chemotherapy. In this study, we investigated possible interactions between ALKBH2 and ALKBH3 with other proteins that could alter damage response and discovered an interaction with the mismatch repair (MMR) system. To test whether the lack of active MMR impacts ALKBH2 and/or ALKBH3 response to methylating agents, we generated cells deficient in ALKBH2, ALKBH3, or both in addition to Mlh homolog 1 (MLH1), another MMR protein. We found that MLH1koALKBH3ko cells showed enhanced resistance toward SN1- and SN2-type methylating agents, whereas MLH1koALKBH2ko cells were only resistant to SN1-type methylating agents. Concomitant loss of ALKBH2 and ALKBH3 (ALKBH2ko3ko) rendered cells sensitive to SN1- and SN2-agents, but the additional loss of MLH1 enhanced resistance to both types of damage. We also showed that ALKBH2ko3ko cells have an ATR-dependent arrest at the G2/M checkpoint, increased apoptotic signaling, and replication fork stress in response to methylation. However, these responses were not observed with the loss of functional MLH1 in MLH1koALKBH2ko3ko cells. Finally, in MLH1koALKBH2ko3ko cells, we observed elevated mutant frequency in untreated and temozolomide treated cells. These results suggest that obtaining a more accurate prognosis of chemotherapeutic outcome requires information on the functionality of ALKBH2, ALKBH3, and MLH1.
Keywords: AlkB; MLH1; demethylation; methylating agents; mismatch repair.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





Similar articles
-
Alkbh2 protects against lethality and mutation in primary mouse embryonic fibroblasts.DNA Repair (Amst). 2012 May 1;11(5):502-10. doi: 10.1016/j.dnarep.2012.02.005. Epub 2012 Mar 17. DNA Repair (Amst). 2012. PMID: 22429847 Free PMC article.
-
CpG promoter methylation of the ALKBH3 alkylation repair gene in breast cancer.BMC Cancer. 2017 Jul 5;17(1):469. doi: 10.1186/s12885-017-3453-8. BMC Cancer. 2017. PMID: 28679371 Free PMC article.
-
DNA repair enzymes ALKBH2, ALKBH3, and AlkB oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in vitro.Nucleic Acids Res. 2019 Jun 20;47(11):5522-5529. doi: 10.1093/nar/gkz395. Nucleic Acids Res. 2019. PMID: 31114894 Free PMC article.
-
Single-stranded DNA damage: Protecting the single-stranded DNA from chemical attack.DNA Repair (Amst). 2020 Mar;87:102804. doi: 10.1016/j.dnarep.2020.102804. Epub 2020 Jan 20. DNA Repair (Amst). 2020. PMID: 31981739 Review.
-
Signalling cell cycle arrest and cell death through the MMR System.Carcinogenesis. 2006 Apr;27(4):682-92. doi: 10.1093/carcin/bgi298. Epub 2005 Dec 6. Carcinogenesis. 2006. PMID: 16332722 Review.
References
-
- Warwick G.P. The mechanism of action of alkylating agents. Cancer Res. 1963;23:1315–1333. - PubMed
-
- Scharer O.D., Jiricny J. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays. 2001;23:270–281. - PubMed
-
- Seeberg E., Eide L., Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. - PubMed
-
- Falnes P.O., Johansen R.F., Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

