Recognition of Chlamydia trachomatis by Toll-like receptor 9 is altered during persistence
- PMID: 38899879
- PMCID: PMC11238561
- DOI: 10.1128/iai.00063-24
Recognition of Chlamydia trachomatis by Toll-like receptor 9 is altered during persistence
Abstract
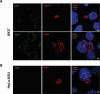
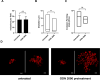
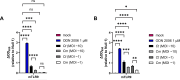
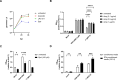
Toll-like receptor 9 (TLR9) is an innate immune receptor that localizes to endosomes in antigen presenting cells and recognizes single stranded unmethylated CpG sites on bacterial genomic DNA (gDNA). Previous bioinformatic studies have demonstrated that the genome of the human pathogen Chlamydia trachomatis contains TLR9 stimulatory motifs, and correlative studies have implied a link between human TLR9 (hTLR9) genotype variants and susceptibility to infection. Here, we present our evaluation of the stimulatory potential of C. trachomatis gDNA and its recognition by hTLR9- and murine TLR9 (mTLR9)-expressing cells. Utilizing reporter cell lines, we demonstrate that purified gDNA from C. trachomatis can stimulate hTLR9 signaling, albeit at lower levels than gDNA prepared from other Gram-negative bacteria. Interestingly, we found that while C. trachomatis is capable of signaling through hTLR9 and mTLR9 during live infections in HEK293 reporter cell lines, signaling only occurs at later developmental time points. Chlamydia-specific induction of hTLR9 is blocked when protein synthesis is inhibited prior to the RB-to-EB conversion, exacerbated by the inhibition of lipooligosaccharide biosynthesis, and is significantly altered during the induction of aberrance/persistence. Our observations support the hypothesis that chlamydial gDNA is released during the conversion between the pathogen's replicative and infectious forms and during treatment with antibiotics targeting peptidoglycan assembly. Given that C. trachomatis inclusions do not co-localize with TLR9-containing vacuoles in the pro-monocytic cell line U937, our findings also hint that chlamydial gDNA is capable of egress from the inclusion, and traffics to TLR9-containing vacuoles via an as yet unknown pathway.
Keywords: Chlamydia; TLR9; innate immunity; pathoadaptation; persistence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

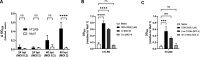
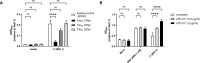
Update of
-
Recognition of Chlamydia trachomatis by Toll-Like Receptor 9 is altered during persistence.bioRxiv [Preprint]. 2024 Feb 8:2024.02.06.579186. doi: 10.1101/2024.02.06.579186. bioRxiv. 2024. Update in: Infect Immun. 2024 Jul 11;92(7):e0006324. doi: 10.1128/iai.00063-24 PMID: 38370826 Free PMC article. Updated. Preprint.
Similar articles
-
Recognition of Chlamydia trachomatis by Toll-Like Receptor 9 is altered during persistence.bioRxiv [Preprint]. 2024 Feb 8:2024.02.06.579186. doi: 10.1101/2024.02.06.579186. bioRxiv. 2024. Update in: Infect Immun. 2024 Jul 11;92(7):e0006324. doi: 10.1128/iai.00063-24 PMID: 38370826 Free PMC article. Updated. Preprint.
-
TLR9 KO mice, haplotypes and CPG indices in Chlamydia trachomatis infection.Drugs Today (Barc). 2009 Nov;45 Suppl B:83-93. Drugs Today (Barc). 2009. PMID: 20011699
-
Selectivity of Human TLR9 for Double CpG Motifs and Implications for the Recognition of Genomic DNA.J Immunol. 2017 Mar 1;198(5):2093-2104. doi: 10.4049/jimmunol.1600757. Epub 2017 Jan 23. J Immunol. 2017. PMID: 28115525
-
Role of toll-like receptors in immune responses to chlamydial infections.Curr Pharm Des. 2008;14(6):593-600. doi: 10.2174/138161208783885344. Curr Pharm Des. 2008. PMID: 18336303 Review.
-
Pathogenic Puppetry: Manipulation of the Host Actin Cytoskeleton by Chlamydia trachomatis.Int J Mol Sci. 2019 Dec 21;21(1):90. doi: 10.3390/ijms21010090. Int J Mol Sci. 2019. PMID: 31877733 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

