This is a preprint.
Human TSC2 Mutant Cells Exhibit Aberrations in Early Neurodevelopment Accompanied by Changes in the DNA Methylome
- PMID: 38895266
- PMCID: PMC11185654
- DOI: 10.1101/2024.06.04.597443
Human TSC2 Mutant Cells Exhibit Aberrations in Early Neurodevelopment Accompanied by Changes in the DNA Methylome
Update in
-
Human TSC2 mutant cells exhibit aberrations in early neurodevelopment accompanied by changes in the DNA Methylome.Hum Mol Genet. 2025 Jan 29:ddae199. doi: 10.1093/hmg/ddae199. Online ahead of print. Hum Mol Genet. 2025. PMID: 39877967
Abstract
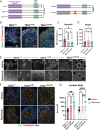
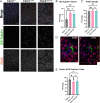
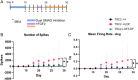
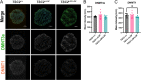
Tuberous Sclerosis Complex (TSC) is a debilitating developmental disorder characterized by a variety of clinical manifestations. While benign tumors in the heart, lungs, kidney, and brain are all hallmarks of the disease, the most severe symptoms of TSC are often neurological, including seizures, autism, psychiatric disorders, and intellectual disabilities. TSC is caused by loss of function mutations in the TSC1 or TSC2 genes and consequent dysregulation of signaling via mechanistic Target of Rapamycin Complex 1 (mTORC1). While TSC neurological phenotypes are well-documented, it is not yet known how early in neural development TSC1/2-mutant cells diverge from the typical developmental trajectory. Another outstanding question is the contribution of homozygous-mutant cells to disease phenotypes and whether such phenotypes are also seen in the heterozygous-mutant populations that comprise the vast majority of cells in patients. Using TSC patient-derived isogenic induced pluripotent stem cells (iPSCs) with defined genetic changes, we observed aberrant early neurodevelopment in vitro, including misexpression of key proteins associated with lineage commitment and premature electrical activity. These alterations in differentiation were coincident with hundreds of differentially methylated DNA regions, including loci associated with key genes in neurodevelopment. Collectively, these data suggest that mutation or loss of TSC2 affects gene regulation and expression at earlier timepoints than previously appreciated, with implications for whether and how prenatal treatment should be pursued.
Figures


Similar articles
-
Human TSC2 mutant cells exhibit aberrations in early neurodevelopment accompanied by changes in the DNA Methylome.Hum Mol Genet. 2025 Jan 29:ddae199. doi: 10.1093/hmg/ddae199. Online ahead of print. Hum Mol Genet. 2025. PMID: 39877967
-
Tuberous Sclerosis Complex.1999 Jul 13 [updated 2024 Aug 1]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 1999 Jul 13 [updated 2024 Aug 1]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301399 Free Books & Documents. Review.
-
Dysregulation of Neurite Outgrowth and Cell Migration in Autism and Other Neurodevelopmental Disorders.Adv Neurobiol. 2020;25:109-153. doi: 10.1007/978-3-030-45493-7_5. Adv Neurobiol. 2020. PMID: 32578146
-
Beckwith-Wiedemann Syndrome.2000 Mar 3 [updated 2023 Sep 21]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2000 Mar 3 [updated 2023 Sep 21]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301568 Free Books & Documents. Review.
-
Genetics, X-Linked Inheritance.2023 May 1. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 1. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 32491315 Free Books & Documents.
References
-
- Crino P.B., Nathanson K.L., and Henske E.P., The tuberous sclerosis complex. N Engl J Med, 2006. 355(13): p. 1345–56. - PubMed
-
- Cardamone M., et al. Mammalian target of rapamycin inhibitors for intractable epilepsy and subependymal giant cell astrocytomas in tuberous sclerosis complex. J Pediatr, 2014. 164(5): p. 1195–200. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
