MATR3's Role beyond the Nuclear Matrix: From Gene Regulation to Its Implications in Amyotrophic Lateral Sclerosis and Other Diseases
- PMID: 38891112
- PMCID: PMC11171696
- DOI: 10.3390/cells13110980
MATR3's Role beyond the Nuclear Matrix: From Gene Regulation to Its Implications in Amyotrophic Lateral Sclerosis and Other Diseases
Abstract
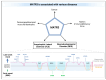
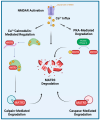
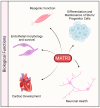
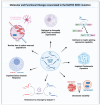
Matrin-3 (MATR3) was initially discovered as a component of the nuclear matrix about thirty years ago. Since then, accumulating studies have provided evidence that MATR3 not only plays a structural role in the nucleus, but that it is also an active protein involved in regulating gene expression at multiple levels, including chromatin organization, DNA transcription, RNA metabolism, and protein translation in the nucleus and cytoplasm. Furthermore, MATR3 may play a critical role in various cellular processes, including DNA damage response, cell proliferation, differentiation, and survival. In addition to the revelation of its biological role, recent studies have reported MATR3's involvement in the context of various diseases, including neurodegenerative and neurodevelopmental diseases, as well as cancer. Moreover, sequencing studies of patients revealed a handful of disease-associated mutations in MATR3 linked to amyotrophic lateral sclerosis (ALS), which further elevated the gene's importance as a topic of study. In this review, we synthesize the current knowledge regarding the diverse functions of MATR3 in DNA- and RNA-related processes, as well as its involvement in various diseases, with a particular emphasis on ALS.
Keywords: DNA-binding protein; MATR3; RNA-binding protein; amyotrophic lateral sclerosis; nucleic acid binding protein.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Matrin 3-dependent neurotoxicity is modified by nucleic acid binding and nucleocytoplasmic localization.Elife. 2018 Jul 17;7:e35977. doi: 10.7554/eLife.35977. Elife. 2018. PMID: 30015619 Free PMC article.
-
RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3.Acta Neuropathol Commun. 2020 Oct 31;8(1):177. doi: 10.1186/s40478-020-01060-y. Acta Neuropathol Commun. 2020. PMID: 33129345 Free PMC article.
-
Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis.Nat Neurosci. 2014 May;17(5):664-666. doi: 10.1038/nn.3688. Epub 2014 Mar 30. Nat Neurosci. 2014. PMID: 24686783 Free PMC article.
-
Matrin 3 in neuromuscular disease: physiology and pathophysiology.JCI Insight. 2021 Jan 11;6(1):e143948. doi: 10.1172/jci.insight.143948. JCI Insight. 2021. PMID: 33427209 Free PMC article. Review.
-
The role of Matrin-3 in physiology and its dysregulation in disease.Biochem Soc Trans. 2024 Jun 26;52(3):961-972. doi: 10.1042/BST20220585. Biochem Soc Trans. 2024. PMID: 38813817 Free PMC article. Review.
Cited by
-
Knockout of Dectin-1 does not modify disease onset or progression in a MATR3 S85C knock-in mouse model of ALS.Heliyon. 2024 Sep 14;10(18):e37926. doi: 10.1016/j.heliyon.2024.e37926. eCollection 2024 Sep 30. Heliyon. 2024. PMID: 39323783 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

