Integrated profiling identifies DXS253E as a potential prognostic marker in colorectal cancer
- PMID: 38890691
- PMCID: PMC11186088
- DOI: 10.1186/s12935-024-03403-4
Integrated profiling identifies DXS253E as a potential prognostic marker in colorectal cancer
Abstract
Background: Increasing evidence suggests that DXS253E is critical for cancer development and progression, but the function and potential mechanism of DXS253E in colorectal cancer (CRC) remain largely unknown. In this study, we evaluated the clinical significance and explored the underlying mechanism of DXS253E in CRC.
Methods: DXS253E expression in cancer tissues was investigated using the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. The Kaplan-Meier plot was used to assess the prognosis of DXS253E. The cBioPortal, MethSurv, and Tumor Immune Estimation Resource (TIMER) databases were employed to analyze the mutation profile, methylation, and immune infiltration associated with DXS253E. The biological functions of DXS253E in CRC cells were determined by CCK-8 assay, plate cloning assay, Transwell assay, flow cytometry, lactate assay, western blot, and qRT-PCR.
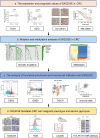
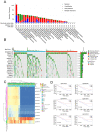
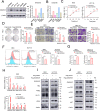
Results: DXS253E was upregulated in CRC tissues and high DXS253E expression levels were correlated with poor survival in CRC patients. Our bioinformatics analyses showed that high DXS253E gene methylation levels were associated with the favorable prognosis of CRC patients. Furthermore, DXS253E levels were linked to the expression levels of several immunomodulatory genes and an abundance of immune cells. Mechanistically, the overexpression of DXS253E enhanced proliferation, migration, invasion, and the aerobic glycolysis of CRC cells through the AKT/mTOR pathway.
Conclusions: We demonstrated that DXS253E functions as a potential role in CRC progression and may serve as an indicator of outcomes and a therapeutic target for regulating the AKT/mTOR pathway in CRC.
Keywords: Bioinformatics; Colorectal cancer; DXS253E; Glycolysis; Prognosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Colorectal cancer with low SLC35A3 is associated with immune infiltrates and poor prognosis.Sci Rep. 2024 Jan 3;14(1):329. doi: 10.1038/s41598-023-51028-w. Sci Rep. 2024. PMID: 38172565 Free PMC article.
-
SEMA6B Overexpression Predicts Poor Prognosis and Correlates With the Tumor Immunosuppressive Microenvironment in Colorectal Cancer.Front Mol Biosci. 2021 Dec 6;8:687319. doi: 10.3389/fmolb.2021.687319. eCollection 2021. Front Mol Biosci. 2021. PMID: 34938771 Free PMC article.
-
Down-regulation of long non-coding RNA RP11-708H21.4 is associated with poor prognosis for colorectal cancer and promotes tumorigenesis through regulating AKT/mTOR pathway.Oncotarget. 2017 Apr 25;8(17):27929-27942. doi: 10.18632/oncotarget.15846. Oncotarget. 2017. PMID: 28427191 Free PMC article.
-
LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF-β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1.Int J Med Sci. 2019 Jan 1;16(1):51-59. doi: 10.7150/ijms.27359. eCollection 2019. Int J Med Sci. 2019. PMID: 30662328 Free PMC article.
-
KIFC3 Promotes Proliferation, Migration, and Invasion in Colorectal Cancer via PI3K/AKT/mTOR Signaling Pathway.Front Genet. 2022 Jun 22;13:848926. doi: 10.3389/fgene.2022.848926. eCollection 2022. Front Genet. 2022. PMID: 35812733 Free PMC article.
References
Grants and funding
- GZC20230142/Program for National Postdoctoral Researcher of China
- 82173218/National Natural Science Foundation of China
- 82171720/National Natural Science Foundation of China
- No. 5202008/Natural Science Foundation of Beijing Municipality
- ZYLX202116/Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support
LinkOut - more resources
Full Text Sources
Miscellaneous

