Deconvoluting Monomer- and Dimer-Specific Distance Distributions between Spin Labels in a Monomer/Dimer Mixture Using T1-Edited DEER EPR Spectroscopy
- PMID: 38888555
- PMCID: PMC11345870
- DOI: 10.1021/jacs.4c03916
Deconvoluting Monomer- and Dimer-Specific Distance Distributions between Spin Labels in a Monomer/Dimer Mixture Using T1-Edited DEER EPR Spectroscopy
Abstract
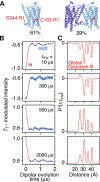
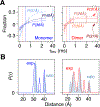
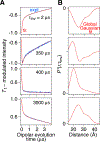
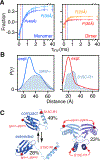
Double electron-electron resonance (DEER) EPR is a powerful tool in structural biology, providing distances between pairs of spin labels. When the sample consists of a mixture of oligomeric species (e.g., monomer and dimer), the question arises as to how to assign the peaks in the DEER-derived probability distance distribution to the individual species. Here, we propose incorporating an EPR longitudinal electron relaxation (T1) inversion recovery experiment within a DEER pulse sequence to resolve this problem. The apparent T1 between dipolar coupled electron spins measured from the inversion recovery time (τinv) dependence of the peak intensities in the T1-edited DEER-derived probability P(r) distance distribution will be affected by the number of nitroxide labels attached to the biomolecule of interest, for example, two for a monomer and four for a dimer. We show that global fitting of all the T1-edited DEER echo curves, recorded over a range of τinv values, permits the deconvolution of distances between spin labels originating from monomeric (longer T1) and dimeric (shorter T1) species. This is especially useful when the trapping of spin labels in different conformational states during freezing gives rise to complex P(r) distance distributions. The utility of this approach is demonstrated for two systems, the β1 adrenergic receptor and a construct of the huntingtin exon-1 protein fused to the immunoglobulin domain of protein G, both of which exist in a monomer-dimer equilibrium.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Quantitative Resolution of Monomer-Dimer Populations by Inversion Modulated DEER EPR Spectroscopy.Chemphyschem. 2016 Oct 5;17(19):2987-2991. doi: 10.1002/cphc.201600726. Epub 2016 Aug 2. Chemphyschem. 2016. PMID: 27442455 Free PMC article.
-
Quantitative analysis of sterol-modulated monomer-dimer equilibrium of the β1-adrenergic receptor by DEER spectroscopy.Proc Natl Acad Sci U S A. 2023 Feb 14;120(7):e2221036120. doi: 10.1073/pnas.2221036120. Epub 2023 Feb 6. Proc Natl Acad Sci U S A. 2023. PMID: 36745787 Free PMC article.
-
Enhancing sensitivity of Double Electron-Electron Resonance (DEER) by using Relaxation-Optimized Acquisition Length Distribution (RELOAD) scheme.J Magn Reson. 2019 Jan;298:115-126. doi: 10.1016/j.jmr.2018.12.004. Epub 2018 Dec 5. J Magn Reson. 2019. PMID: 30544015 Free PMC article.
-
Nitroxide spin labels and EPR spectroscopy: A powerful association for protein dynamics studies.Biochim Biophys Acta Proteins Proteom. 2021 Jul;1869(7):140653. doi: 10.1016/j.bbapap.2021.140653. Epub 2021 Mar 20. Biochim Biophys Acta Proteins Proteom. 2021. PMID: 33757896 Review.
-
Exploring protein conformations in vitro and in cell with EPR distance measurements.Curr Opin Struct Biol. 2022 Aug;75:102398. doi: 10.1016/j.sbi.2022.102398. Epub 2022 Jun 3. Curr Opin Struct Biol. 2022. PMID: 35667279 Review.
References
-
- Schiemann O; Prisner TF Long-Range Distance Determinations in Biomacromolecules by EPR Spectroscopy. Q. Rev. Biophys 2007, 40, 1–53 - PubMed
-
- Jeschke G Deer Distance Measurements on Proteins. Annu. Rev. Phys. Chem 2012, 63, 419–446. - PubMed
-
- Bode BE; Margraf D; Plackmeyer J; Durner G; Prisner TF; Schiemann O Counting the Monomers in Nanometer-Sized Oligomers by Pulsed Electron-Electron Double Resonance. J. Am. Chem. Soc 2007, 129, 6736–6745. - PubMed
-
- Jeschke G; Sajid M; Schulte M; Godt A Three-Spin Correlations in Double Electron-Electron Resonance. Phys. Chem. Chem. Phys 2009, 11, 6580–6591. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

