Comprehensive bioinformatics analysis of human cytomegalovirus pathway genes in pan-cancer
- PMID: 38886862
- PMCID: PMC11181644
- DOI: 10.1186/s40246-024-00633-5
Comprehensive bioinformatics analysis of human cytomegalovirus pathway genes in pan-cancer
Abstract
Background: Human cytomegalovirus (HCMV) is a herpesvirus that can infect various cell types and modulate host gene expression and immune response. It has been associated with the pathogenesis of various cancers, but its molecular mechanisms remain elusive.
Methods: We comprehensively analyzed the expression of HCMV pathway genes across 26 cancer types using the Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx) databases. We also used bioinformatics tools to study immune invasion and tumor microenvironment in pan-cancer. Cox regression and machine learning were used to analyze prognostic genes and their relationship with drug sensitivity.
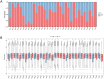
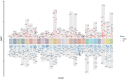
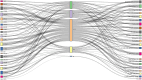
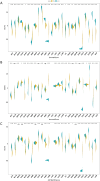
Results: We found that HCMV pathway genes are widely expressed in various cancers. Immune infiltration and the tumor microenvironment revealed that HCMV is involved in complex immune processes. We obtained prognostic genes for 25 cancers and significantly found 23 key genes in the HCMV pathway, which are significantly enriched in cellular chemotaxis and synaptic function and may be involved in disease progression. Notably, CaM family genes were up-regulated and AC family genes were down-regulated in most tumors. These hub genes correlate with sensitivity or resistance to various drugs, suggesting their potential as therapeutic targets.
Conclusions: Our study has revealed the role of the HCMV pathway in various cancers and provided insights into its molecular mechanism and therapeutic significance. It is worth noting that the key genes of the HCMV pathway may open up new doors for cancer prevention and treatment.
Keywords: Bioinformatics; Human cytomegalovirus pathway; Immune infiltration; Pan-cancer; Tumor mutation burden.
© 2024. The Author(s).
Conflict of interest statement
All the authors declared that they had no competing interests.
Figures

Similar articles
-
Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA.Gene. 2014 Feb 25;536(2):272-8. doi: 10.1016/j.gene.2013.12.012. Epub 2013 Dec 18. Gene. 2014. PMID: 24361963
-
TCGA Database-Based Screening of Tumor Microenvironment Immunomodulators Related to Bladder Cancer Prognosis.Ann Clin Lab Sci. 2024 May;54(3):299-312. Ann Clin Lab Sci. 2024. PMID: 39048164
-
Human cytomegalovirus infection enhances cell proliferation, migration and upregulation of EMT markers in colorectal cancer-derived stem cell-like cells.Int J Oncol. 2017 Nov;51(5):1415-1426. doi: 10.3892/ijo.2017.4135. Epub 2017 Sep 25. Int J Oncol. 2017. PMID: 29048611 Free PMC article.
-
Human cytomegalovirus (HCMV)-encoded microRNAs: potential biomarkers and clinical applications.RNA Biol. 2021 Dec;18(12):2194-2202. doi: 10.1080/15476286.2021.1930757. Epub 2021 May 26. RNA Biol. 2021. PMID: 34039247 Free PMC article. Review.
-
Immune Landscape of CMV Infection in Cancer Patients: From "Canonical" Diseases Toward Virus-Elicited Oncomodulation.Front Immunol. 2021 Sep 8;12:730765. doi: 10.3389/fimmu.2021.730765. eCollection 2021. Front Immunol. 2021. PMID: 34566995 Free PMC article. Review.
References
-
- Sung H, Global Cancer Statistics. 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71(3):209-249.10.3322/caac.21660. - PubMed
Publication types
MeSH terms
Grants and funding
- 82160537/The National Natural Science Foundation of China
- 2022A1515011368/The Natural Science Foundation of Guangdong Province of China
- 2022ZDZX2057/The Key Projects of Department of Education of Guangdong Province of China
- Guike AB22035027/Guangxi Key Research and Development Project
- 2023YFC2605400/National Key Research and Development Program of China
LinkOut - more resources
Full Text Sources

