This is a preprint.
Key features of the innate immune response is mediated by the immunoproteasome in microglia
- PMID: 38883799
- PMCID: PMC11177974
- DOI: 10.21203/rs.3.rs-4467983/v1
Key features of the innate immune response is mediated by the immunoproteasome in microglia
Abstract
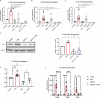
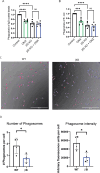
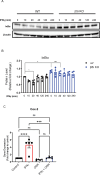
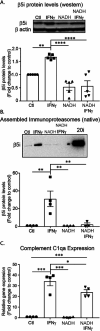
Microglia are the resident immune cells of the central nervous system (CNS). We and others have shown that the inflammatory response of microglia is partially regulated by the immunoproteasome, an inducible form of the proteasome responsible for the generation of major histocompatibility complex (MHC) class I epitopes. While the role of the proteasome in the adaptive immune system is well established, emerging evidence suggests the immunoproteasome may have discrete functions in the innate immune response. Here, we show that inhibiting the immunoproteasome reduces the IFNγ-dependent induction of complement activator C1q, suppresses phagocytosis, and alters the cytokine expression profile in a microglial cell line and microglia derived from human inducible pluripotent stem cells. Moreover, we show that the immunoproteasome regulates the degradation of IκBα, a modulator of NF-κB signaling. Finally, we demonstrate that NADH prevents induction of the immunoproteasome, representing a potential pathway to suppress immunoproteasome-dependent immune responses.
Keywords: Complement; Cytokines; Immunoproteasome; Innate immunity; Microglia; NFκ-B; ONX-0914; Phagocytosis.
Conflict of interest statement
Neither I nor my family members have a financial interest in any commercial product, service, or organization providing financial support for this research.
Figures

Similar articles
-
The role of the immunoproteasome in interferon-γ-mediated microglial activation.Sci Rep. 2017 Aug 24;7(1):9365. doi: 10.1038/s41598-017-09715-y. Sci Rep. 2017. PMID: 28839214 Free PMC article.
-
Immunoproteasomes control activation of innate immune signaling and microglial function.Front Immunol. 2022 Oct 6;13:982786. doi: 10.3389/fimmu.2022.982786. eCollection 2022. Front Immunol. 2022. PMID: 36275769 Free PMC article.
-
Proteasome function shapes innate and adaptive immune responses.Am J Physiol Lung Cell Mol Physiol. 2016 Aug 1;311(2):L328-36. doi: 10.1152/ajplung.00156.2016. Epub 2016 Jun 24. Am J Physiol Lung Cell Mol Physiol. 2016. PMID: 27343191 Review.
-
Acetaldehyde suppresses the display of HBV-MHC class I complexes on HBV-expressing hepatocytes.Am J Physiol Gastrointest Liver Physiol. 2019 Aug 1;317(2):G127-G140. doi: 10.1152/ajpgi.00064.2019. Epub 2019 May 29. Am J Physiol Gastrointest Liver Physiol. 2019. PMID: 31141391 Free PMC article.
-
The immunoproteasome and viral infection: a complex regulator of inflammation.Front Microbiol. 2015 Jan 29;6:21. doi: 10.3389/fmicb.2015.00021. eCollection 2015. Front Microbiol. 2015. PMID: 25688236 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

