No evidence for ongoing replication on ART in SIV-infected macaques
- PMID: 38877003
- PMCID: PMC11178840
- DOI: 10.1038/s41467-024-49369-9
No evidence for ongoing replication on ART in SIV-infected macaques
Abstract
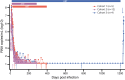
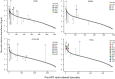
The capacity of HIV-1 to replicate during optimal antiretroviral therapy (ART) is challenging to assess directly. To gain greater sensitivity to detect evolution on ART, we used a nonhuman primate (NHP) model providing precise control over the level of pre-ART evolution and more comprehensive analyses than are possible with clinical samples. We infected 21 rhesus macaques (RMs) with the barcoded virus SIVmac239M and initiated ART early to minimize baseline genetic diversity. RMs were treated for 285-1200 days. We used several tests of molecular evolution to compare 1352 near-full-length (nFL) SIV DNA single genome sequences from PBMCs, lymph nodes, and spleen obtained near the time of ART initiation and those present after long-term ART, none of which showed significant changes to the SIV DNA population during ART in any animal. To investigate the possibility of ongoing replication in unsampled putative tissue sanctuaries during ART, we discontinued treatment in four animals and confirmed that none of the 336 nFL SIV RNA sequences obtained from rebound plasma viremia showed evidence of evolution. The rigorous nature of our analyses reinforced the emerging consensus of a lack of appreciable ongoing replication on effective ART and validates the relevance of this NHP model for cure studies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Evaluating the Intactness of Persistent Viral Genomes in Simian Immunodeficiency Virus-Infected Rhesus Macaques after Initiating Antiretroviral Therapy within One Year of Infection.J Virol. 2019 Dec 12;94(1):e01308-19. doi: 10.1128/JVI.01308-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31597776 Free PMC article.
-
SMAC Mimetic Plus Triple-Combination Bispecific HIVxCD3 Retargeting Molecules in SHIV.C.CH505-Infected, Antiretroviral Therapy-Suppressed Rhesus Macaques.J Virol. 2020 Oct 14;94(21):e00793-20. doi: 10.1128/JVI.00793-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32817214 Free PMC article.
-
Short-Term Pegylated Interferon α2a Treatment Does Not Significantly Reduce the Viral Reservoir of Simian Immunodeficiency Virus-Infected, Antiretroviral Therapy-Treated Rhesus Macaques.J Virol. 2018 Jun 29;92(14):e00279-18. doi: 10.1128/JVI.00279-18. Print 2018 Jul 15. J Virol. 2018. PMID: 29720521 Free PMC article.
-
Virologic and Immunologic Features of Simian Immunodeficiency Virus Control Post-ART Interruption in Rhesus Macaques.J Virol. 2020 Jul 1;94(14):e00338-20. doi: 10.1128/JVI.00338-20. Print 2020 Jul 1. J Virol. 2020. PMID: 32350073 Free PMC article.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

