Characterization of Cwc2, U6 snRNA, and Prp8 interactions destabilized by Prp16 ATPase at the transition between the first and second steps of splicing
- PMID: 38876504
- PMCID: PMC11331412
- DOI: 10.1261/rna.079886.123
Characterization of Cwc2, U6 snRNA, and Prp8 interactions destabilized by Prp16 ATPase at the transition between the first and second steps of splicing
Abstract
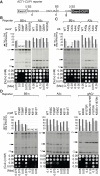
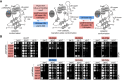
The spliceosome performs two consecutive transesterification reactions using one catalytic center, thus requiring its rearrangement between the two catalytic steps of splicing. The Prp16 ATPase facilitates exit from the first-step conformation of the catalytic center by destabilizing some interactions important for catalysis. To better understand rearrangements within the Saccharomyces cerevisiae catalytic center, we characterize factors that modulate the function of Prp16: Cwc2, N-terminal domain of Prp8, and U6-41AACAAU46 region. Alleles of these factors were identified through genetic screens for mutants that correct cs defects of prp16-302 alleles. Several of the identified U6, cwc2, and prp8 alleles are located in close proximity of each other in cryo-EM structures of the spliceosomal catalytic conformations. Cwc2 and U6 interact with the intron sequences in the first step, but they do not seem to contribute to the stability of the second-step catalytic center. On the other hand, the N-terminal segment of Prp8 not only affects intron positioning for the first step, but it also makes important contacts in the proximity of the active site for both the first and second steps of splicing. By identifying interactions important for the stability of catalytic conformations, our genetic analyses indirectly inform us about features of the transition-state conformation of the spliceosome.
Keywords: Prp16; Prp8; U6 snRNA; spliceosomal catalytic center; transition-state conformation.
© 2024 Meissner et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures






Similar articles
-
Structure of a spliceosome remodelled for exon ligation.Nature. 2017 Feb 16;542(7641):377-380. doi: 10.1038/nature21078. Epub 2017 Jan 11. Nature. 2017. PMID: 28076345 Free PMC article.
-
Functional analysis of the zinc finger modules of the Saccharomyces cerevisiae splicing factor Luc7.RNA. 2024 Jul 16;30(8):1058-1069. doi: 10.1261/rna.079956.124. RNA. 2024. PMID: 38719745
-
Structure and function of an RNase H domain at the heart of the spliceosome.EMBO J. 2008 Nov 5;27(21):2929-40. doi: 10.1038/emboj.2008.209. Epub 2008 Oct 9. EMBO J. 2008. PMID: 18843295 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Pharmacological treatments in panic disorder in adults: a network meta-analysis.Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
