This is a preprint.
Multi-omics profiling of mouse polycystic kidney disease progression at a single cell resolution
- PMID: 38854144
- PMCID: PMC11160654
- DOI: 10.1101/2024.05.27.595830
Multi-omics profiling of mouse polycystic kidney disease progression at a single cell resolution
Update in
-
Multiomics profiling of mouse polycystic kidney disease progression at a single-cell resolution.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2410830121. doi: 10.1073/pnas.2410830121. Epub 2024 Oct 15. Proc Natl Acad Sci U S A. 2024. PMID: 39405347
Abstract
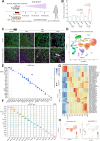
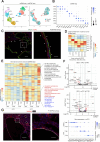
Autosomal dominant polycystic kidney disease (ADPKD) is the most common hereditary kidney disease and causes significant morbidity, ultimately leading to end-stage kidney disease. PKD pathogenesis is characterized by complex and dynamic alterations in multiple cell types during disease progression, hampering a deeper understanding of disease mechanism and the development of therapeutic approaches. Here, we generate a single nucleus multimodal atlas of an orthologous mouse PKD model at early, mid and late timepoints, consisting of 125,434 single-nucleus transcriptomic and epigenetic multiomes. We catalogue differentially expressed genes and activated epigenetic regions in each cell type during PKD progression, characterizing cell-type-specific responses to Pkd1 deletion. We describe heterogeneous, atypical collecting duct cells as well as proximal tubular cells that constitute cyst epithelia in PKD. The transcriptional regulation of the cyst lining cell marker GPRC5A is conserved between mouse and human PKD cystic epithelia, suggesting shared gene regulatory pathways. Our single nucleus multiomic analysis of mouse PKD provides a foundation to understand the earliest changes molecular deregulation in a mouse model of PKD at a single-cell resolution.
Conflict of interest statement
Competing Interest Statement: B.D.H. is a consultant for Janssen Research & Development, LLC, Pfizer and Chinook Therapeutics, holds equity in Chinook Therapeutics and grant funding from Chinook Therapeutics and Janssen Research & Development, LLC. O.M.W has received grants from AstraZeneca unrelated to the current work. The remaining authors declare no competing interests.
Figures



Similar articles
-
Multiomics profiling of mouse polycystic kidney disease progression at a single-cell resolution.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2410830121. doi: 10.1073/pnas.2410830121. Epub 2024 Oct 15. Proc Natl Acad Sci U S A. 2024. PMID: 39405347
-
Pkd2 Deficiency in Embryonic Aqp2 + Progenitor Cells Is Sufficient to Cause Severe Polycystic Kidney Disease.J Am Soc Nephrol. 2024 Apr 1;35(4):398-409. doi: 10.1681/ASN.0000000000000309. Epub 2024 Jan 23. J Am Soc Nephrol. 2024. PMID: 38254271
-
Defining cellular complexity in human autosomal dominant polycystic kidney disease by multimodal single cell analysis.Nat Commun. 2022 Oct 30;13(1):6497. doi: 10.1038/s41467-022-34255-z. Nat Commun. 2022. PMID: 36310237 Free PMC article.
-
The master regulators Myc and p53 cellular signaling and functions in polycystic kidney disease.Cell Signal. 2020 Jul;71:109594. doi: 10.1016/j.cellsig.2020.109594. Epub 2020 Mar 4. Cell Signal. 2020. PMID: 32145315 Review.
-
Molecular Mechanisms of Epigenetic Regulation, Inflammation, and Cell Death in ADPKD.Front Mol Biosci. 2022 Jun 29;9:922428. doi: 10.3389/fmolb.2022.922428. eCollection 2022. Front Mol Biosci. 2022. PMID: 35847973 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
