Riboseq-flow: A streamlined, reliable pipeline for ribosome profiling data analysis and quality control
- PMID: 38846930
- PMCID: PMC11153996
- DOI: 10.12688/wellcomeopenres.21000.1
Riboseq-flow: A streamlined, reliable pipeline for ribosome profiling data analysis and quality control
Abstract
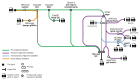
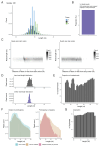
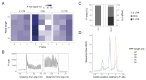
Ribosome profiling is a powerful technique to study translation at a transcriptome-wide level. However, ensuring good data quality is paramount for accurate interpretation, as is ensuring that the analyses are reproducible. We introduce a new Nextflow DSL2 pipeline, riboseq-flow, designed for processing and comprehensive quality control of ribosome profiling experiments. Riboseq-flow is user-friendly, versatile and upholds high standards in reproducibility, scalability, portability, version control and continuous integration. It enables users to efficiently analyse multiple samples in parallel and helps them evaluate the quality and utility of their data based on the detailed metrics and visualisations that are automatically generated. Riboseq-flow is available at https://github.com/iraiosub/riboseq-flow.
Keywords: Nextflow; ribo-seq; ribosome profiling.
Plain language summary
Ribosome profiling is a cutting-edge method that provides a detailed view of protein synthesis across the entire set of RNA molecules within cells. To ensure the reliability of such studies, high-quality data and the ability to replicate analyses are crucial. To address this, we present riboseq-flow, a new tool built with Nextflow DSL2, tailored for analysing data from ribosome profiling experiments. This pipeline stands out for its ease of use, flexibility, and commitment to high reproducibility standards. It's designed to handle multiple samples simultaneously, ensuring efficient analysis for large-scale studies. Moreover, riboseq-flow automatically generates detailed reports and visual representations to assess the data quality, enhancing researchers' understanding of their experiments and guiding future decisions. This valuable resource is freely accessible at https://github.com/iraiosub/riboseq-flow.
Copyright: © 2024 Iosub IA et al.
Conflict of interest statement
No competing interests were disclosed.
Figures

Similar articles
-
RiboDoc: A Docker-based package for ribosome profiling analysis.Comput Struct Biotechnol J. 2021 May 7;19:2851-2860. doi: 10.1016/j.csbj.2021.05.014. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34093996 Free PMC article.
-
RiboGalaxy: A Galaxy-based Web Platform for Ribosome Profiling Data Processing - 2023 Update.J Mol Biol. 2023 Jul 15;435(14):168043. doi: 10.1016/j.jmb.2023.168043. Epub 2023 Mar 9. J Mol Biol. 2023. PMID: 37356899
-
riboviz: analysis and visualization of ribosome profiling datasets.BMC Bioinformatics. 2017 Oct 25;18(1):461. doi: 10.1186/s12859-017-1873-8. BMC Bioinformatics. 2017. PMID: 29070028 Free PMC article.
-
Insights into the mechanisms of eukaryotic translation gained with ribosome profiling.Nucleic Acids Res. 2017 Jan 25;45(2):513-526. doi: 10.1093/nar/gkw1190. Epub 2016 Dec 6. Nucleic Acids Res. 2017. PMID: 27923997 Free PMC article. Review.
-
Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale.Wiley Interdiscip Rev RNA. 2013 Sep-Oct;4(5):473-90. doi: 10.1002/wrna.1172. Epub 2013 May 20. Wiley Interdiscip Rev RNA. 2013. PMID: 23696005 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

