Single-cell and spatial transcriptomics reveal a high glycolysis B cell and tumor-associated macrophages cluster correlated with poor prognosis and exhausted immune microenvironment in diffuse large B-cell lymphoma
- PMID: 38840205
- PMCID: PMC11155084
- DOI: 10.1186/s40364-024-00605-w
Single-cell and spatial transcriptomics reveal a high glycolysis B cell and tumor-associated macrophages cluster correlated with poor prognosis and exhausted immune microenvironment in diffuse large B-cell lymphoma
Abstract
Background: Diffuse large B-cell lymphoma (DLBCL) is a heterogeneous malignancy characterized by varied responses to treatment and prognoses. Understanding the metabolic characteristics driving DLBCL progression is crucial for developing personalized therapies.
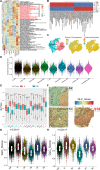
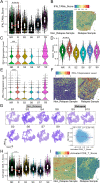
Methods: This study utilized multiple omics technologies including single-cell transcriptomics (n = 5), bulk transcriptomics (n = 966), spatial transcriptomics (n = 10), immunohistochemistry (n = 34), multiple immunofluorescence (n = 20) and to elucidate the metabolic features of highly malignant DLBCL cells and tumor-associated macrophages (TAMs), along with their associated tumor microenvironment. Metabolic pathway analysis facilitated by scMetabolism, and integrated analysis via hdWGCNA, identified glycolysis genes correlating with malignancy, and the prognostic value of glycolysis genes (STMN1, ENO1, PKM, and CDK1) and TAMs were verified.
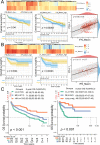
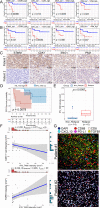
Results: High-glycolysis malignant DLBCL tissues exhibited an immunosuppressive microenvironment characterized by abundant IFN_TAMs (CD68+CXCL10+PD-L1+) and diminished CD8+ T cell infiltration. Glycolysis genes were positively correlated with malignancy degree. IFN_TAMs exhibited high glycolysis activity and closely communicating with high-malignancy DLBCL cells identified within datasets. The glycolysis score, evaluated by seven genes, emerged as an independent prognostic factor (HR = 1.796, 95% CI: 1.077-2.995, p = 0.025 and HR = 2.631, 95% CI: 1.207-5.735, p = 0.015) along with IFN_TAMs were positively correlated with poor survival (p < 0.05) in DLBCL. Immunohistochemical validation of glycolysis markers (STMN1, ENO1, PKM, and CDK1) and multiple immunofluorescence validation of IFN_TAMs underscored their prognostic value (p < 0.05) in DLBCL.
Conclusions: This study underscores the significance of glycolysis in tumor progression and modulation of the immune microenvironment. The identified glycolysis genes and IFN_TAMs represent potential prognostic markers and therapeutic targets in DLBCL.
Keywords: Diffuse large B-cell lymphoma; Glycolysis; Metabolism; Single-cell transcriptomics; Spatial transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors report there are no competing interests to declare. All authors agreed to submit for consideration for publication in this journal.
Figures





Similar articles
-
High PTX3 expression is associated with a poor prognosis in diffuse large B-cell lymphoma.Cancer Sci. 2022 Jan;113(1):334-348. doi: 10.1111/cas.15179. Epub 2021 Nov 29. Cancer Sci. 2022. PMID: 34706126 Free PMC article.
-
Characterisation of tumour microenvironment and immune checkpoints in primary central nervous system diffuse large B cell lymphomas.Virchows Arch. 2020 Jun;476(6):891-902. doi: 10.1007/s00428-019-02695-6. Epub 2019 Dec 6. Virchows Arch. 2020. PMID: 31811434
-
Prognostic implications of the tumor immune microenvironment and immune checkpoint pathway in primary central nervous system diffuse large B-cell lymphoma in the North Indian population.APMIS. 2022 Feb;130(2):82-94. doi: 10.1111/apm.13195. Epub 2021 Dec 23. APMIS. 2022. PMID: 34862664
-
Prognostic and clinicopathological significance of PD-1/PD-L1 expression in the tumor microenvironment and neoplastic cells for lymphoma.Int Immunopharmacol. 2019 Dec;77:105999. doi: 10.1016/j.intimp.2019.105999. Epub 2019 Nov 6. Int Immunopharmacol. 2019. PMID: 31704289 Review.
-
Unraveling the Immune Microenvironment in Diffuse Large B-Cell Lymphoma: Prognostic and Potential Therapeutic Implications.Curr Issues Mol Biol. 2024 Jul 5;46(7):7048-7064. doi: 10.3390/cimb46070420. Curr Issues Mol Biol. 2024. PMID: 39057061 Free PMC article. Review.
Cited by
-
Tumor Biology Hides Novel Therapeutic Approaches to Diffuse Large B-Cell Lymphoma: A Narrative Review.Int J Mol Sci. 2024 Oct 23;25(21):11384. doi: 10.3390/ijms252111384. Int J Mol Sci. 2024. PMID: 39518937 Free PMC article. Review.
-
Metabolic reprogramming and therapeutic resistance in primary and metastatic breast cancer.Mol Cancer. 2024 Nov 21;23(1):261. doi: 10.1186/s12943-024-02165-x. Mol Cancer. 2024. PMID: 39574178 Free PMC article. Review.
References
Grants and funding
- No. 2023A1515110952/Guangdong Basic and Applied Basic Research Foundation
- CICAMS-MOMP2022006/Major Project of Medical Oncology Key Foundation of Cancer Hospital Chinese Academy of Medical Sciences
- CIFMS 2021-I2M-1-003/CAMS Innovation Fund for Medical Sciences
- 2022-PUMCH-B-033/National High Level Hospital Clinical Research Funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

