Nanoparticle delivery of Tat synergizes with classical latency reversal agents to express HIV antigen targets
- PMID: 38829049
- PMCID: PMC11232404
- DOI: 10.1128/aac.00201-24
Nanoparticle delivery of Tat synergizes with classical latency reversal agents to express HIV antigen targets
Abstract
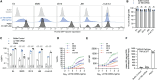
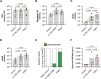
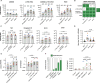
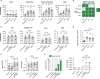
Limited cellular levels of the HIV transcriptional activator Tat are one contributor to proviral latency that might be targeted in HIV cure strategies. We recently demonstrated that lipid nanoparticles containing HIV tat mRNA induce HIV expression in primary CD4 T cells. Here, we sought to further characterize tat mRNA in the context of several benchmark latency reversal agents (LRAs), including inhibitor of apoptosis protein antagonists (IAPi), bromodomain and extra-Terminal motif inhibitors (BETi), and histone deacetylase inhibitors (HDACi). tat mRNA reversed latency across several different cell line models of HIV latency, an effect dependent on the TAR hairpin loop. Synergistic enhancement of tat mRNA activity was observed with IAPi, HDACi, and BETi, albeit to variable degrees. In primary CD4 T cells from durably suppressed people with HIV, tat mRNA profoundly increased the frequencies of elongated, multiply-spliced, and polyadenylated HIV transcripts, while having a lesser impact on TAR transcript frequencies. tat mRNAs alone resulted in variable HIV p24 protein induction across donors. However, tat mRNA in combination with IAPi, BETi, or HDACi markedly enhanced HIV RNA and protein expression without overt cytotoxicity or cellular activation. Notably, combination regimens approached or in some cases exceeded the latency reversal activity of maximal mitogenic T cell stimulation. Higher levels of tat mRNA-driven HIV p24 induction were observed in donors with larger mitogen-inducible HIV reservoirs, and expression increased with prolonged exposure time. Combination LRA strategies employing both small molecule inhibitors and Tat delivered to CD4 T cells are a promising approach to effectively target the HIV reservoir.
Keywords: HIV; Tat; latency; lipid; nanoparticles; reversal.
Conflict of interest statement
J.V. was an employee of Arcturus and may be a stockholder. E.V.G. and D.B. were employees of Johnson & Johnson and may be stockholders. D.M.M. has consulted for Merck and ViiV, outside the area of this work, and holds common stock in Gilead. No other conflicts are declared.
Figures

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
HIV-1 latency reversal agent boosting is not limited by opioid use.JCI Insight. 2024 Nov 22;9(22):e185480. doi: 10.1172/jci.insight.185480. JCI Insight. 2024. PMID: 39470739 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
"I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article.
-
Methods of milk expression for lactating women.Cochrane Database Syst Rev. 2016 Sep 29;9(9):CD006170. doi: 10.1002/14651858.CD006170.pub5. Cochrane Database Syst Rev. 2016. PMID: 27684560 Free PMC article. Review.
References
-
- Li JZ, Aga E, Bosch RJ, Pilkinton M, Kroon E, MacLaren L, Keefer M, Fox L, Barr L, Acosta E, Ananworanich J, Coombs R, Mellors JW, Landay AL, Macatangay B, Deeks S, Gandhi RT, Smith DM. 2022. Time to viral rebound after interruption of modern antiretroviral therapies. Clin Infect Dis 74:865–870. doi:10.1093/cid/ciab541 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- UM1-AI164567/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U54-AI170792/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- UM1 AI164567/AI/NIAID NIH HHS/United States
- U54 AI170792/AI/NIAID NIH HHS/United States
- P30 AI050410/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

