This is a preprint.
Viral and host network analysis of the human cytomegalovirus transcriptome in latency
- PMID: 38826434
- PMCID: PMC11142044
- DOI: 10.1101/2024.05.21.594597
Viral and host network analysis of the human cytomegalovirus transcriptome in latency
Abstract
HCMV genes UL135 and UL138 play opposing roles regulating latency and reactivation in CD34+ human progenitor cells (HPCs). Using the THP-1 cell line model for latency and reactivation, we designed an RNA sequencing study to compare the transcriptional profile of HCMV infection in the presence and absence of these genes. The loss of UL138 results in elevated levels of viral gene expression and increased differentiation of cell populations that support HCMV gene expression and genome synthesis. The loss of UL135 results in diminished viral gene expression during an initial burst that occurs as latency is established and no expression of eleven viral genes from the ULb' region even following stimulation for differentiation and reactivation. Transcriptional network analysis revealed host transcription factors with potential to regulate the ULb' genes in coordination with pUL135. These results reveal roles for UL135 and UL138 in regulation of viral gene expression and potentially hematopoietic differentiation.
Conflict of interest statement
Declaration of Interests: The authors declare no competing interests.
Figures

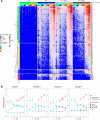
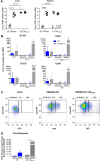
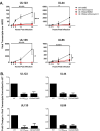
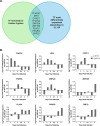
Similar articles
-
Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection.J Virol. 2014 Jun;88(11):5987-6002. doi: 10.1128/JVI.03506-13. Epub 2014 Mar 12. J Virol. 2014. PMID: 24623432 Free PMC article.
-
Human Cytomegalovirus UL135 Interacts with Host Adaptor Proteins To Regulate Epidermal Growth Factor Receptor and Reactivation from Latency.J Virol. 2018 Sep 26;92(20):e00919-18. doi: 10.1128/JVI.00919-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30089695 Free PMC article.
-
Stabilization of the human cytomegalovirus UL136p33 reactivation determinant overcomes the requirement for UL135 for replication in hematopoietic cells.J Virol. 2023 Aug 31;97(8):e0014823. doi: 10.1128/jvi.00148-23. Epub 2023 Aug 11. J Virol. 2023. PMID: 37565749 Free PMC article.
-
The Role of the Human Cytomegalovirus UL133-UL138 Gene Locus in Latency and Reactivation.Viruses. 2020 Jul 1;12(7):714. doi: 10.3390/v12070714. Viruses. 2020. PMID: 32630219 Free PMC article. Review.
-
Cell signaling and cytomegalovirus reactivation: what do Src family kinases have to do with it?Biochem Soc Trans. 2020 Apr 29;48(2):667-675. doi: 10.1042/BST20191110. Biochem Soc Trans. 2020. PMID: 32311019 Free PMC article. Review.
References
-
- Goodrum F., Britt W., Mocarski E. S., in Fields Virology, Howley P. M., Knipe D. M., Damania B. A., Cohen J. I., Eds. (Lippincott Williams & Wilkins, Philadelphia, PA, 2021), vol. 2, chap. 12, pp. 389–444.
-
- Chambers J., Angulo A., Amaratunga D., Guo H., Jiang Y., Wan J. S., Bittner A., Frueh K., Jackson M. R., Peterson P. A., Erlander M. G., Ghazal P., DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. Journal of virology 73, 5757–5766 (1999); published online EpubJul (10.1128/jvi.73.7.5757-5766.1999). - DOI - PMC - PubMed
-
- Gatherer D., Seirafian S., Cunningham C., Holton M., Dargan D. J., Baluchova K., Hector R. D., Galbraith J., Herzyk P., Wilkinson G. W., Davison A. J., High-resolution human cytomegalovirus transcriptome. Proc Natl Acad Sci U S A 108, 19755–19760 (2011); published online EpubDec 6 (10.1073/pnas.1115861108). - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
