Effect of folA gene in human breast milk-derived Limosilactobacillus reuteri on its folate biosynthesis
- PMID: 38812695
- PMCID: PMC11133606
- DOI: 10.3389/fmicb.2024.1402654
Effect of folA gene in human breast milk-derived Limosilactobacillus reuteri on its folate biosynthesis
Abstract
Introduction: Folate supplementation is crucial for the human body, and the chemically synthesized folic acid might have undesirable side effects. The use of molecular breeding methods to modify the genes related to the biosynthesis of folate by probiotics to increase folate production is currently a focus of research.
Methods: In this study, the folate-producing strain of Limosilactobacillus reuteri B1-28 was isolated from human breast milk, and the difference between B1-28 and folA gene deletion strain ΔFolA was investigated by phenotyping, in vitro probiotic evaluation, metabolism and transcriptome analysis.
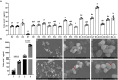
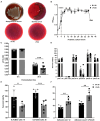
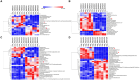
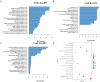
Results: The results showed that the folate producted by the ΔFolA was 2-3 folds that of the B1-28. Scanning electron microscope showed that ΔFolA had rougher surface, and the acid-producing capacity (p = 0.0008) and adhesion properties (p = 0.0096) were significantly enhanced than B1-28. Transcriptomic analysis revealed that differentially expressed genes were mainly involved in three pathways, among which the biosynthesis of ribosome and aminoacyl-tRNA occurred in the key metabolic pathways. Metabolomics analysis showed that folA affected 5 metabolic pathways, involving 89 different metabolites.
Discussion: In conclusion, the editing of a key gene of folA in folate biosynthesis pathway provides a feasible pathway to improve folate biosynthesis in breast milk-derived probiotics.
Keywords: Limosilactobacillus reuteri; folate; metabolism; molecular breeding; probiotic properties; transcription.
Copyright © 2024 Jiang, Li, Zhang, Ji, Yang, Liu, Zhang, Qiao, Zhao, Du, Fan, Dang, Chen, Jiang and Chen.
Conflict of interest statement
YuJ, XL, WZ, YaJ, KY, LL, MZ, WQ, JZ, MD, XF, XD, HC, and LC were employed by the Beijing Sanyuan Foods Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features.PLoS One. 2011 Apr 29;6(4):e18783. doi: 10.1371/journal.pone.0018783. PLoS One. 2011. PMID: 21559529 Free PMC article.
-
In Silico Genomic and Metabolic Atlas of Limosilactobacillus reuteri DSM 20016: An Insight into Human Health.Microorganisms. 2022 Jul 2;10(7):1341. doi: 10.3390/microorganisms10071341. Microorganisms. 2022. PMID: 35889060 Free PMC article.
-
The probiotic and immunomodulation effects of Limosilactobacillus reuteri RGW1 isolated from calf feces.Front Cell Infect Microbiol. 2023 Jan 12;12:1086861. doi: 10.3389/fcimb.2022.1086861. eCollection 2022. Front Cell Infect Microbiol. 2023. PMID: 36710979 Free PMC article.
-
Microbiota and Probiotics: The Role of Limosilactobacillus Reuteri in Diverticulitis.Medicina (Kaunas). 2021 Aug 5;57(8):802. doi: 10.3390/medicina57080802. Medicina (Kaunas). 2021. PMID: 34441008 Free PMC article. Review.
-
Lactobacillus Reuteri DSM 17938 (Limosilactobacillus reuteri) in Diarrhea and Constipation: Two Sides of the Same Coin?Medicina (Kaunas). 2021 Jun 23;57(7):643. doi: 10.3390/medicina57070643. Medicina (Kaunas). 2021. PMID: 34201542 Free PMC article. Review.
References
-
- Chavez-Esquivel G., Cervantes-Cuevas H., Ybieta-Olvera L. F., Castañeda Briones M. T., Acosta D., Cabello J. (2021). Antimicrobial activity of graphite oxide doped with silver against Bacillus subtilis, Candida albicans, Escherichia coli, and Staphylococcus aureus by agar well diffusion test: synthesis and characterization. Mater. Sci. Eng. C Mater. Biol. Appl. 123:111934. doi: 10.1016/j.msec.2021.111934 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

