CRISPR Screen Reveals PACT as a Pro-Viral Factor for Dengue Viral Replication
- PMID: 38793607
- PMCID: PMC11125577
- DOI: 10.3390/v16050725
CRISPR Screen Reveals PACT as a Pro-Viral Factor for Dengue Viral Replication
Abstract
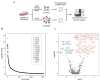
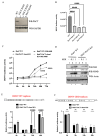
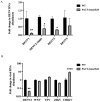
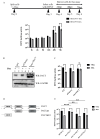
The dengue virus is a single-stranded, positive-sense RNA virus that infects ~400 million people worldwide. Currently, there are no approved antivirals available. CRISPR-based screening methods have greatly accelerated the discovery of host factors that are essential for DENV infection and that can be targeted in host-directed antiviral interventions. In the present study, we performed a focused CRISPR (Clustered Regularly Interspaced Palindromic Repeats) library screen to discover the key host factors that are essential for DENV infection in human Huh7 cells and identified the Protein Activator of Interferon-Induced Protein Kinase (PACT) as a novel pro-viral factor for DENV. PACT is a double-stranded RNA-binding protein generally known to activate antiviral responses in virus-infected cells and block viral replication. However, in our studies, we observed that PACT plays a pro-viral role in DENV infection and specifically promotes viral RNA replication. Knockout of PACT resulted in a significant decrease in DENV RNA and protein abundances in infected cells, which was rescued upon ectopic expression of full-length PACT. An analysis of global gene expression changes indicated that several ER-associated pro-viral genes such as ERN1, DDIT3, HERPUD1, and EIF2AK3 are not upregulated in DENV-infected PACT knockout cells as compared to infected wildtype cells. Thus, our study demonstrates a novel role for PACT in promoting DENV replication, possibly through modulating the expression of ER-associated pro-viral genes.
Keywords: CRISPR screen; dengue virus; host–virus interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A Genome-Wide CRISPR-Cas9 Screen Identifies the Dolichol-Phosphate Mannose Synthase Complex as a Host Dependency Factor for Dengue Virus Infection.J Virol. 2020 Mar 17;94(7):e01751-19. doi: 10.1128/JVI.01751-19. Print 2020 Mar 17. J Virol. 2020. PMID: 31915280 Free PMC article.
-
Identification of RNA Binding Proteins Associated with Dengue Virus RNA in Infected Cells Reveals Temporally Distinct Host Factor Requirements.PLoS Negl Trop Dis. 2016 Aug 24;10(8):e0004921. doi: 10.1371/journal.pntd.0004921. eCollection 2016 Aug. PLoS Negl Trop Dis. 2016. PMID: 27556644 Free PMC article.
-
Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens.Nature. 2016 Jul 7;535(7610):159-63. doi: 10.1038/nature18631. Epub 2016 Jun 17. Nature. 2016. PMID: 27383987 Free PMC article.
-
The Transactions of NS3 and NS5 in Flaviviral RNA Replication.Adv Exp Med Biol. 2018;1062:147-163. doi: 10.1007/978-981-10-8727-1_11. Adv Exp Med Biol. 2018. PMID: 29845531 Review.
-
How Dengue Virus Circumvents Innate Immunity.Front Immunol. 2018 Dec 4;9:2860. doi: 10.3389/fimmu.2018.02860. eCollection 2018. Front Immunol. 2018. PMID: 30564245 Free PMC article. Review.
Cited by
-
Exploring the Frontiers of Virus-Host Interactions-3rd Edition.Viruses. 2024 Sep 30;16(10):1544. doi: 10.3390/v16101544. Viruses. 2024. PMID: 39459878 Free PMC article.
-
A review of virus host factor discovery using CRISPR screening.mBio. 2024 Nov 13;15(11):e0320523. doi: 10.1128/mbio.03205-23. Epub 2024 Oct 18. mBio. 2024. PMID: 39422472 Free PMC article. Review.
References
-
- Labeau A., Simon-Loriere E., Hafirassou M.L., Bonnet-Madin L., Tessier S., Zamborlini A., Dupre T., Seta N., Schwartz O., Chaix M.L., et al. A Genome-Wide CRISPR-Cas9 Screen Identifies the Dolichol-Phosphate Mannose Synthase Complex as a Host Dependency Factor for Dengue Virus Infection. J. Virol. 2020;94:e01751-19. doi: 10.1128/JVI.01751-19. - DOI - PMC - PubMed
-
- Savidis G., McDougall W.M., Meraner P., Perreira J.M., Portmann J.M., Trincucci G., John S.P., Aker A.M., Renzette N., Robbins D.R., et al. Identification of Zika Virus and Dengue Virus Dependency Factors using Functional Genomics. Cell Rep. 2016;16:232–246. doi: 10.1016/j.celrep.2016.06.028. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- R01 AI141970/AI/NIAID NIH HHS/United States
- JNC/2023/02064/Jawaharlal Nehru Centre for Advanced Scientific Research
- R01 AI069000 (to P.S.), R01 AI140186 (to J.E.C.); NIH R01 AI141970 (to J.E.C.); NIH R01 AI130123 (to J.E.C.)/NH/NIH HHS/United States
- SERB/2023/01976/Department of Science and Technology
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

