A Combined Molecular Dynamics and Hydropathic INTeraction (HINT) Approach to Investigate Protein Flexibility: The PPARγ Case Study
- PMID: 38792097
- PMCID: PMC11124508
- DOI: 10.3390/molecules29102234
A Combined Molecular Dynamics and Hydropathic INTeraction (HINT) Approach to Investigate Protein Flexibility: The PPARγ Case Study
Abstract
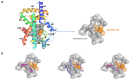
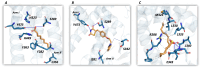
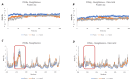
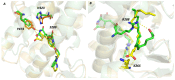
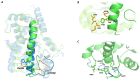
Molecular Dynamics (MD) is a computational technique widely used to evaluate a molecular system's thermodynamic properties and conformational behavior over time. In particular, the energy analysis of a protein conformation ensemble produced though MD simulations plays a crucial role in explaining the relationship between protein dynamics and its mechanism of action. In this research work, the HINT (Hydropathic INTeractions) LogP-based scoring function was first used to handle MD trajectories and investigate the molecular basis behind the intricate PPARγ mechanism of activation. The Peroxisome Proliferator-Activated Receptor γ (PPARγ) is an emblematic example of a highly flexible protein due to the extended ω-loop delimiting the active site, and it is responsible for the receptor's ability to bind chemically different compounds. In this work, we focused on the PPARγ complex with Rosiglitazone, a common anti-diabetic compound and analyzed the molecular basis of the flexible ω-loop stabilization effect produced by the Oleic Acid co-binding. The HINT-based analysis of the produced MD trajectories allowed us to account for all of the energetic contributions involved in interconverting between conformational states and describe the intramolecular interactions between the flexible ω-loop and the helix H3 triggered by the allosteric binding mechanism.
Keywords: HINT force field; Molecular Dynamics; PPARγ; conformational analysis; mechanism of action.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Microsecond MD Simulations to Explore the Structural and Energetic Differences between the Human RXRα-PPARγ vs. RXRα-PPARγ-DNA.Molecules. 2022 Sep 7;27(18):5778. doi: 10.3390/molecules27185778. Molecules. 2022. PMID: 36144514 Free PMC article.
-
A computational study to identify the key residues of peroxisome proliferator-activated receptor gamma in the interactions with its antagonists.J Biomol Struct Dyn. 2018 May;36(7):1822-1833. doi: 10.1080/07391102.2017.1335618. Epub 2017 Jun 14. J Biomol Struct Dyn. 2018. PMID: 28566016
-
Molecular Modeling Approach to Study the PPARγ-Ligand Interactions.Methods Mol Biol. 2019;1966:261-289. doi: 10.1007/978-1-4939-9195-2_22. Methods Mol Biol. 2019. PMID: 31041755
-
Peroxisome Proliferator-Activated Receptor γ (PPARγ) and Ligand Choreography: Newcomers Take the Stage.J Med Chem. 2015 Jul 23;58(14):5381-94. doi: 10.1021/jm501155f. Epub 2015 Apr 9. J Med Chem. 2015. PMID: 25734377 Review.
-
Insights into Dynamic Mechanism of Ligand Binding to Peroxisome Proliferator-Activated Receptor γ toward Potential Pharmacological Applications.Biol Pharm Bull. 2021;44(9):1185-1195. doi: 10.1248/bpb.b21-00263. Biol Pharm Bull. 2021. PMID: 34471046 Review.
References
-
- Gund P., Halgren T.A., Smith G.M. Annual Reports in Medicinal Chemistry. Academic Press; Cambridge, MA, USA: 1987. Chapter 27 Molecular Modeling as an Aid to Drug Design and Discovery; pp. 269–279. - DOI
-
- Tripathi M.K., Ahmad S., Tyagi R., Dahiya V., Yadav M.K. Computer Aided Drug Design (CADD): From Ligand-Based Methods to Structure-Based Approaches. Elsevier; Amsterdam, The Netherlands: 2022. Fundamentals of molecular modeling in drug design; pp. 125–155. - DOI
-
- Adelusi T.I., Oyedele A.-Q.K., Boyenle I.D., Ogunlana A.T., Adeyemi R.O., Ukachi C.D., Idris M.O., Olaoba O.T., Adedotun I.O., Kolawole O.E., et al. Molecular modeling in drug discovery. Inform. Med. Unlocked. 2022;29:100880. doi: 10.1016/j.imu.2022.100880. - DOI
-
- van Gunsteren W.F., Berendsen H.J.C. Computer Simulation of Molecular Dynamics: Methodology, Applications, and Perspectives in Chemistry. Angew. Chem. Int. Ed. 1990;29:992–1023. doi: 10.1002/anie.199009921. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

